Scrutinizing virus genome termini by high-throughput sequencing
- PMID: 24465717
- PMCID: PMC3896407
- DOI: 10.1371/journal.pone.0085806
Scrutinizing virus genome termini by high-throughput sequencing
Abstract
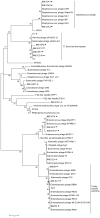
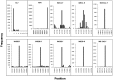
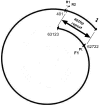
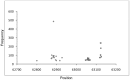
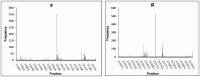
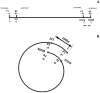
Analysis of genomic terminal sequences has been a major step in studies on viral DNA replication and packaging mechanisms. However, traditional methods to study genome termini are challenging due to the time-consuming protocols and their inefficiency where critical details are lost easily. Recent advances in next generation sequencing (NGS) have enabled it to be a powerful tool to study genome termini. In this study, using NGS we sequenced one iridovirus genome and twenty phage genomes and confirmed for the first time that the high frequency sequences (HFSs) found in the NGS reads are indeed the terminal sequences of viral genomes. Further, we established a criterion to distinguish the type of termini and the viral packaging mode. We also obtained additional terminal details such as terminal repeats, multi-termini, asymmetric termini. With this approach, we were able to simultaneously detect details of the genome termini as well as obtain the complete sequence of bacteriophage genomes. Theoretically, this application can be further extended to analyze larger and more complicated genomes of plant and animal viruses. This study proposed a novel and efficient method for research on viral replication, packaging, terminase activity, transcription regulation, and metabolism of the host cell.
Conflict of interest statement
Figures

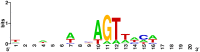

Similar articles
-
Predicting genome terminus sequences of Bacillus cereus-group bacteriophage using next generation sequencing data.BMC Genomics. 2017 May 4;18(1):350. doi: 10.1186/s12864-017-3744-0. BMC Genomics. 2017. PMID: 28472946 Free PMC article.
-
Analyzing Genome Termini of Bacteriophage Through High-Throughput Sequencing.Methods Mol Biol. 2018;1681:139-163. doi: 10.1007/978-1-4939-7343-9_11. Methods Mol Biol. 2018. PMID: 29134593
-
Sequence characteristics of T4-like bacteriophage IME08 benome termini revealed by high throughput sequencing.Virol J. 2011 Apr 27;8:194. doi: 10.1186/1743-422X-8-194. Virol J. 2011. PMID: 21524290 Free PMC article.
-
Next-Generation Sequencing in the Understanding of Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Biology.Viruses. 2016 Mar 31;8(4):92. doi: 10.3390/v8040092. Viruses. 2016. PMID: 27043613 Free PMC article. Review.
-
Next-generation sequencing technology in clinical virology.Clin Microbiol Infect. 2013 Jan;19(1):15-22. doi: 10.1111/1469-0691.12056. Clin Microbiol Infect. 2013. PMID: 23279287 Review.
Cited by
-
Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies.BMC Genomics. 2016 Aug 26;17(1):679. doi: 10.1186/s12864-016-3018-2. BMC Genomics. 2016. PMID: 27561606 Free PMC article.
-
PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data.Sci Rep. 2017 Aug 15;7(1):8292. doi: 10.1038/s41598-017-07910-5. Sci Rep. 2017. PMID: 28811656 Free PMC article.
-
Morganella Phage Mecenats66 Utilizes an Evolutionarily Distinct Subtype of Headful Genome Packaging with a Preferred Packaging Initiation Site.Microorganisms. 2022 Sep 7;10(9):1799. doi: 10.3390/microorganisms10091799. Microorganisms. 2022. PMID: 36144401 Free PMC article.
-
The large terminase DNA packaging motor grips DNA with its ATPase domain for cleavage by the flexible nuclease domain.Nucleic Acids Res. 2017 Apr 7;45(6):3591-3605. doi: 10.1093/nar/gkw1356. Nucleic Acids Res. 2017. PMID: 28082398 Free PMC article.
-
Pectobacterium atrosepticum Phage vB_PatP_CB5: A Member of the Proposed Genus 'Phimunavirus'.Viruses. 2018 Jul 26;10(8):394. doi: 10.3390/v10080394. Viruses. 2018. PMID: 30050020 Free PMC article.
References
-
- Kutter E, Sulakvelidze A (2005) Bacteriophages: biology and applications: CRC.
-
- Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annual review of genetics 42: 647–681. - PubMed
-
- Fujisawa H, Morita M (1997) Phage DNA packaging. Genes to Cells 2: 537–545. - PubMed
-
- Catalano CE, Cue D, Feiss M (1995) Virus DNA packaging: the strategy used by phage λ. Molecular microbiology 16: 1075–1086. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

