The intracellular bacteria Chlamydia hijack peroxisomes and utilize their enzymatic capacity to produce bacteria-specific phospholipids
- PMID: 24465954
- PMCID: PMC3900481
- DOI: 10.1371/journal.pone.0086196
The intracellular bacteria Chlamydia hijack peroxisomes and utilize their enzymatic capacity to produce bacteria-specific phospholipids
Abstract
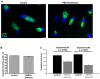
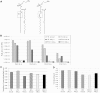
Chlamydia trachomatis is an obligate intracellular pathogen responsible for loss of eyesight through trachoma and for millions of cases annually of sexually transmitted diseases. The bacteria develop within a membrane-bounded inclusion. They lack enzymes for several biosynthetic pathways, including those to make some phospholipids, and exploit their host to compensate. Three-dimensional fluorescence microscopy demonstrates that small organelles of the host, peroxisomes, are translocated into the Chlamydia inclusion and are found adjacent to the bacteria. In cells deficient for peroxisome biogenesis the bacteria are able to multiply and give rise to infectious progeny, demonstrating that peroxisomes are not essential for bacterial development in vitro. Mass spectrometry-based lipidomics reveal the presence in C. trachomatis of plasmalogens, ether phospholipids whose synthesis begins in peroxisomes and have never been described in aerobic bacteria before. Some of the bacterial plasmalogens are novel structures containing bacteria-specific odd-chain fatty acids; they are not made in uninfected cells nor in peroxisome-deficient cells. Their biosynthesis is thus accomplished by the metabolic collaboration of peroxisomes and bacteria.
Conflict of interest statement
Figures



Similar articles
-
Host and Bacterial Glycolysis during Chlamydia trachomatis Infection.Infect Immun. 2020 Nov 16;88(12):e00545-20. doi: 10.1128/IAI.00545-20. Print 2020 Nov 16. Infect Immun. 2020. PMID: 32900818 Free PMC article.
-
Chlamydia trachomatis regulates growth and development in response to host cell fatty acid availability in the absence of lipid droplets.Cell Microbiol. 2018 Feb;20(2):10.1111/cmi.12801. doi: 10.1111/cmi.12801. Epub 2017 Dec 12. Cell Microbiol. 2018. PMID: 29117636 Free PMC article.
-
Chlamydia trachomatis Scavenges Host Fatty Acids for Phospholipid Synthesis via an Acyl-Acyl Carrier Protein Synthetase.J Biol Chem. 2015 Sep 4;290(36):22163-73. doi: 10.1074/jbc.M115.671008. Epub 2015 Jul 20. J Biol Chem. 2015. PMID: 26195634 Free PMC article.
-
Plasmalogen homeostasis - regulation of plasmalogen biosynthesis and its physiological consequence in mammals.FEBS Lett. 2017 Sep;591(18):2720-2729. doi: 10.1002/1873-3468.12743. Epub 2017 Jul 28. FEBS Lett. 2017. PMID: 28686302 Review.
-
Imaging of Chlamydia and host cell metabolism.Future Microbiol. 2014;9(4):509-21. doi: 10.2217/fmb.14.13. Future Microbiol. 2014. PMID: 24810350 Review.
Cited by
-
Modulation of Host Lipid Pathways by Pathogenic Intracellular Bacteria.Pathogens. 2020 Jul 28;9(8):614. doi: 10.3390/pathogens9080614. Pathogens. 2020. PMID: 32731350 Free PMC article. Review.
-
The marine bacterium Phaeobacter inhibens secures external ammonium by rapid buildup of intracellular nitrogen stocks.FEMS Microbiol Ecol. 2018 Oct 1;94(10):fiy154. doi: 10.1093/femsec/fiy154. FEMS Microbiol Ecol. 2018. PMID: 30124819 Free PMC article.
-
Phosphatidylserine decarboxylase CT699, lysophospholipid acyltransferase CT775, and acyl-ACP synthase CT776 provide membrane lipid diversity to Chlamydia trachomatis.Sci Rep. 2017 Nov 17;7(1):15767. doi: 10.1038/s41598-017-16116-8. Sci Rep. 2017. PMID: 29150677 Free PMC article.
-
An endometrial organoid model of interactions between Chlamydia and epithelial and immune cells.J Cell Sci. 2021 Mar 8;134(5):jcs252403. doi: 10.1242/jcs.252403. J Cell Sci. 2021. PMID: 33468625 Free PMC article.
-
Chlamydia trachomatis growth depends on eukaryotic cholesterol esterification and is affected by Acyl-CoA:cholesterol acyltransferase inhibition.Pathog Dis. 2015 Aug;73(6):ftv028. doi: 10.1093/femspd/ftv028. Epub 2015 Apr 15. Pathog Dis. 2015. PMID: 25883118 Free PMC article.
References
-
- Geisler WM (2010) Duration of Untreated, Uncomplicated Chlamydia trachomatis Genital Infection and Factors Associated with Chlamydia Resolution: A Review of Human Studies. J Infect Dis 201: S104–S113. - PubMed
-
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, et al. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis . Science 282: 754–755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

