Design and characterization of a 52K SNP chip for goats
- PMID: 24465974
- PMCID: PMC3899236
- DOI: 10.1371/journal.pone.0086227
Design and characterization of a 52K SNP chip for goats
Erratum in
-
Correction: Design and Characterization of a 52K SNP Chip for Goats.PLoS One. 2016 Mar 24;11(3):e0152632. doi: 10.1371/journal.pone.0152632. eCollection 2016. PLoS One. 2016. PMID: 27011020 Free PMC article. No abstract available.
Abstract
The success of Genome Wide Association Studies in the discovery of sequence variation linked to complex traits in humans has increased interest in high throughput SNP genotyping assays in livestock species. Primary goals are QTL detection and genomic selection. The purpose here was design of a 50-60,000 SNP chip for goats. The success of a moderate density SNP assay depends on reliable bioinformatic SNP detection procedures, the technological success rate of the SNP design, even spacing of SNPs on the genome and selection of Minor Allele Frequencies (MAF) suitable to use in diverse breeds. Through the federation of three SNP discovery projects consolidated as the International Goat Genome Consortium, we have identified approximately twelve million high quality SNP variants in the goat genome stored in a database together with their biological and technical characteristics. These SNPs were identified within and between six breeds (meat, milk and mixed): Alpine, Boer, Creole, Katjang, Saanen and Savanna, comprising a total of 97 animals. Whole genome and Reduced Representation Library sequences were aligned on >10 kb scaffolds of the de novo goat genome assembly. The 60,000 selected SNPs, evenly spaced on the goat genome, were submitted for oligo manufacturing (Illumina, Inc) and published in dbSNP along with flanking sequences and map position on goat assemblies (i.e. scaffolds and pseudo-chromosomes), sheep genome V2 and cattle UMD3.1 assembly. Ten breeds were then used to validate the SNP content and 52,295 loci could be successfully genotyped and used to generate a final cluster file. The combined strategy of using mainly whole genome Next Generation Sequencing and mapping on a contig genome assembly, complemented with Illumina design tools proved to be efficient in producing this GoatSNP50 chip. Advances in use of molecular markers are expected to accelerate goat genomic studies in coming years.
Conflict of interest statement
Figures

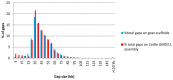
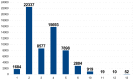
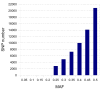
References
-
- Taberlet P, Coissac E, Pansu J, Pompanon F (2011) Conservation genetics of cattle, sheep, and goats. C R Biol 334: 247–254. - PubMed
-
- Schibler L, Vaiman D, Oustry A, Guinec N, Dangy-Caye AL, et al. (1998) Construction and extensive characterization of a goat bacterial artificial chromosome library with threefold genome coverage. Mamm Genome 9: 119–124. - PubMed
-
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP (1998) Comparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res 8: 901–915. - PubMed
-
- Pailhoux E, Vigier B, Chaffaux S, Servel N, Taourit S, et al. (2001) A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat Genet 29: 453–458. - PubMed
-
- Vaiman D, Koutita O, Oustry A, Elsen JM, Manfredi E, et al. (1996) Genetic mapping of the autosomal region involved in XX sex-reversal and horn development in goats. Mamm Genome 7: 133–137. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

