Use of peptide nucleic acids to manipulate gene expression in the malaria parasite Plasmodium falciparum
- PMID: 24466246
- PMCID: PMC3899306
- DOI: 10.1371/journal.pone.0086802
Use of peptide nucleic acids to manipulate gene expression in the malaria parasite Plasmodium falciparum
Abstract
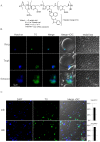
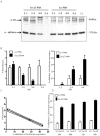
One of the major concerns in treating malaria by conventional small drug molecules is the rapid emergence of drug resistance. Specific silencing of essential genes by antisense oliogomers has been proposed as an alternative approach that may result in antimalarial activity which is not associated with drug resistance. In addition, such an approach could be an important biological tool for studying many genes' function by reverse genetics. Here we present a novel methodology of using peptide nucleic acids (PNAs) as a useful tool for gene silencing in Plasmodium falciparum. PNAs, designed as specific antisense molecules, were conjugated to a cell penetrating peptide (CPP); namely, octa-D-lysine via the C-terminus, to allow facile delivery through cell membranes. PNAs added to P. falciparum cultures were found exclusively in infected erythrocytes and were eventually localized in nuclei of the parasites at all stages of intra erythrocytic development. We show that these PNAs specifically down regulated both a stably expressed transgene as well as an endogenous essential gene, which significantly reduced parasites' viability. This study paves the way for a simple approach to silence a variety of P. falciparum genes as means of deciphering their function and potentially to develop highly specific and potent antimalarial agents.
Conflict of interest statement
Figures

Similar articles
-
Improved cellular uptake of antisense peptide nucleic acids by conjugation to a cell-penetrating peptide and a lipid domain.Methods Mol Biol. 2011;751:209-21. doi: 10.1007/978-1-61779-151-2_13. Methods Mol Biol. 2011. PMID: 21674333
-
Identifying Plasmodium falciparum cytoadherence-linked asexual protein 3 (CLAG 3) sequences that specifically bind to C32 cells and erythrocytes.Protein Sci. 2005 Feb;14(2):504-13. doi: 10.1110/ps.04883905. Protein Sci. 2005. PMID: 15659379 Free PMC article.
-
Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length.Biochemistry. 2001 Jan 9;40(1):53-64. doi: 10.1021/bi0020630. Biochemistry. 2001. PMID: 11141056
-
miRNA therapeutics: delivery and biological activity of peptide nucleic acids targeting miRNAs.Epigenomics. 2011 Dec;3(6):733-45. doi: 10.2217/epi.11.90. Epigenomics. 2011. PMID: 22126292 Review.
-
Developments in Cell-Penetrating Peptides as Antiviral Agents and as Vehicles for Delivery of Peptide Nucleic Acid Targeting Hepadnaviral Replication Pathway.Biomolecules. 2018 Jul 16;8(3):55. doi: 10.3390/biom8030055. Biomolecules. 2018. PMID: 30013006 Free PMC article. Review.
Cited by
-
Polyanionic Carboxyethyl Peptide Nucleic Acids (ce-PNAs): Synthesis and DNA Binding.PLoS One. 2015 Oct 15;10(10):e0140468. doi: 10.1371/journal.pone.0140468. eCollection 2015. PLoS One. 2015. PMID: 26469337 Free PMC article.
-
Exploring the Potential of miRNA-92a-3p as Lead for Sequence-Based Therapies for Malaria.Acta Parasitol. 2025 Aug 11;70(4):179. doi: 10.1007/s11686-025-01117-9. Acta Parasitol. 2025. PMID: 40788442
-
Dual RNA-seq to catalogue host and parasite gene expression changes associated with virulence of T. annulata-transformed bovine leukocytes: towards identification of attenuation biomarkers.Sci Rep. 2023 Oct 24;13(1):18202. doi: 10.1038/s41598-023-45458-9. Sci Rep. 2023. PMID: 37875584 Free PMC article.
-
Targeting protein translation, RNA splicing, and degradation by morpholino-based conjugates in Plasmodium falciparum.Proc Natl Acad Sci U S A. 2015 Sep 22;112(38):11935-40. doi: 10.1073/pnas.1515864112. Epub 2015 Sep 8. Proc Natl Acad Sci U S A. 2015. PMID: 26351679 Free PMC article.
-
Peptide nucleic acids: Advanced tools for biomedical applications.J Biotechnol. 2017 Oct 10;259:148-159. doi: 10.1016/j.jbiotec.2017.07.026. Epub 2017 Jul 29. J Biotechnol. 2017. PMID: 28764969 Free PMC article. Review.
References
-
- Armstrong CM, Goldberg DE (2007) An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods 4: 1007–1009. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

