Tetramer formation in Arabidopsis MADS domain proteins: analysis of a protein-protein interaction network
- PMID: 24468197
- PMCID: PMC3913338
- DOI: 10.1186/1752-0509-8-9
Tetramer formation in Arabidopsis MADS domain proteins: analysis of a protein-protein interaction network
Abstract
Background: MADS domain proteins are transcription factors that coordinate several important developmental processes in plants. These proteins interact with other MADS domain proteins to form dimers, and it has been proposed that they are able to associate as tetrameric complexes that regulate transcription of target genes. Whether the formation of functional tetramers is a widespread property of plant MADS domain proteins, or it is specific to few of these transcriptional regulators remains unclear.
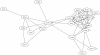
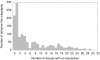
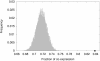
Results: We analyzed the structure of the network of physical interactions among MADS domain proteins in Arabidopsis thaliana. We determined the abundance of subgraphs that represent the connection pattern expected for a MADS domain protein heterotetramer. These subgraphs were significantly more abundant in the MADS domain protein interaction network than in randomized analogous networks. Importantly, these subgraphs are not significantly frequent in a protein interaction network of TCP plant transcription factors, when compared to expectation by chance. In addition, we found that MADS domain proteins in tetramer-like subgraphs are more likely to be expressed jointly than proteins in other subgraphs. This effect is mainly due to proteins in the monophyletic MIKC clade, as there is no association between tetramer-like subgraphs and co-expression for proteins outside this clade.
Conclusions: Our results support that the tendency to form functional tetramers is widespread in the MADS domain protein-protein interaction network. Our observations also suggest that this trend is prevalent, or perhaps exclusive, for proteins in the MIKC clade. Because it is possible to retrodict several experimental results from our analyses, our work can be an important aid to make new predictions and facilitates experimental research on plant MADS domain proteins.
Figures

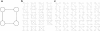
Similar articles
-
A conserved leucine zipper-like motif accounts for strong tetramerization capabilities of SEPALLATA-like MADS-domain transcription factors.J Exp Bot. 2018 Apr 9;69(8):1943-1954. doi: 10.1093/jxb/ery063. J Exp Bot. 2018. PMID: 29474620 Free PMC article.
-
Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis.Plant Cell. 2014 Sep;26(9):3603-15. doi: 10.1105/tpc.114.127910. Epub 2014 Sep 16. Plant Cell. 2014. PMID: 25228343 Free PMC article.
-
MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis.Plant Physiol. 2009 Apr;149(4):1713-23. doi: 10.1104/pp.109.135806. Epub 2009 Feb 11. Plant Physiol. 2009. PMID: 19211705 Free PMC article.
-
MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants.Gene. 2005 Mar 14;347(2):183-98. doi: 10.1016/j.gene.2004.12.014. Epub 2005 Feb 22. Gene. 2005. PMID: 15777618 Review.
-
The emerging importance of type I MADS box transcription factors for plant reproduction.Plant Cell. 2011 Mar;23(3):865-72. doi: 10.1105/tpc.110.081737. Epub 2011 Mar 4. Plant Cell. 2011. PMID: 21378131 Free PMC article. Review.
Cited by
-
Cracking the Floral Quartet Code: How Do Multimers of MIKCC-Type MADS-Domain Transcription Factors Recognize Their Target Genes?Int J Mol Sci. 2023 May 4;24(9):8253. doi: 10.3390/ijms24098253. Int J Mol Sci. 2023. PMID: 37175955 Free PMC article. Review.
-
Identification of rice Di19 family reveals OsDi19-4 involved in drought resistance.Plant Cell Rep. 2014 Dec;33(12):2047-62. doi: 10.1007/s00299-014-1679-3. Epub 2014 Sep 20. Plant Cell Rep. 2014. PMID: 25236158
-
The Origin of Floral Quartet Formation-Ancient Exon Duplications Shaped the Evolution of MIKC-type MADS-domain Transcription Factor Interactions.Mol Biol Evol. 2023 May 2;40(5):msad088. doi: 10.1093/molbev/msad088. Mol Biol Evol. 2023. PMID: 37043523 Free PMC article.
-
Structural Basis for Plant MADS Transcription Factor Oligomerization.Comput Struct Biotechnol J. 2019 Jun 14;17:946-953. doi: 10.1016/j.csbj.2019.06.014. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31360333 Free PMC article. Review.
-
Exploring potential new floral organ morphogenesis genes of Arabidopsis thaliana using systems biology approach.Front Plant Sci. 2015 Oct 13;6:829. doi: 10.3389/fpls.2015.00829. eCollection 2015. Front Plant Sci. 2015. PMID: 26528302 Free PMC article.
References
-
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell. 2003;15(7):1538–1551. doi: 10.1105/tpc.011544. doi:10.1105/tpc.011544.these. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

