A rapid and massive gene expression shift marking adolescent transition in C. elegans
- PMID: 24468752
- PMCID: PMC3904150
- DOI: 10.1038/srep03912
A rapid and massive gene expression shift marking adolescent transition in C. elegans
Abstract
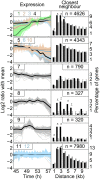
Organismal development is the most dynamic period of the life cycle, yet we have only a rough understanding of the dynamics of gene expression during adolescent transition. Here we show that adolescence in Caenorhabditis elegans is characterized by a spectacular expression shift of conserved and highly polymorphic genes. Using a high resolution time series we found that in adolescent worms over 10,000 genes changed their expression. These genes were clustered according to their expression patterns. One cluster involved in chromatin remodelling showed a brief up-regulation around 50 h post-hatch. At the same time a spectacular shift in expression was observed. Sequence comparisons for this cluster across many genotypes revealed diversifying selection. Strongly up-regulated genes showed signs of purifying selection in non-coding regions, indicating that adolescence-active genes are constrained on their regulatory properties. Our findings improve our understanding of adolescent transition and help to eliminate experimental artefacts due to incorrect developmental timing.
Figures



References
-
- Gutteling E. W., Riksen J. A., Bakker J. & Kammenga J. E. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98, 28–37, 10.1038/sj.hdy.6800894 (2007). - PubMed
-
- Francesconi M. & Lehner B. The effects of genetic variation on gene expression dynamics during development. Nature, 10.1038/nature12772 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

