TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis
- PMID: 24469049
- PMCID: PMC4112181
- DOI: 10.1038/onc.2013.569
TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis
Abstract
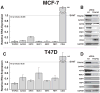
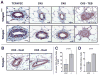
Molecular subtypes of breast cancer are characterized by distinct patterns of gene expression that are predictive of outcome and response to therapy. The luminal breast cancer subtypes are defined by the expression of estrogen receptor-alpha (ERα)-associated genes, many of which are directly responsive to the transcription factor activator protein 2C (TFAP2C). TFAP2C participates in a gene regulatory network controlling cell growth and differentiation during ectodermal development and regulating ESR1/ERα and other luminal cell-associated genes in breast cancer. TFAP2C has been established as a prognostic factor in human breast cancer, however, its role in the establishment and maintenance of the luminal cell phenotype during carcinogenesis and mammary gland development have remained elusive. Herein, we demonstrate a critical role for TFAP2C in maintaining the luminal phenotype in human breast cancer and in influencing the luminal cell phenotype during normal mammary development. Knockdown of TFAP2C in luminal breast carcinoma cells induced epithelial-mesenchymal transition with morphological and phenotypic changes characterized by a loss of luminal-associated gene expression and a concomitant gain of basal-associated gene expression. Conditional knockout of the mouse homolog of TFAP2C, Tcfap2c, in mouse mammary epithelium driven by MMTV-Cre promoted aberrant growth of the mammary tree leading to a reduction in the CD24(hi)/CD49f(mid) luminal cell population and concomitant gain of the CD24(mid)/CD49f(hi) basal cell population at maturity. Our results establish TFAP2C as a key transcriptional regulator for maintaining the luminal phenotype in human breast carcinoma. Furthermore, Tcfap2c influences development of the luminal cell type during mammary development. The data suggest that TFAP2C has an important role in regulated luminal-specific genes and may be a viable therapeutic target in breast cancer.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest.
Figures





References
-
- Friedrichs N, Steiner S, Buettner R, Knoepfle G. Immunohistochemical expression patterns of AP2alpha and AP2gamma in the developing fetal human breast. Histopathology. 2007;51:814–823. - PubMed
-
- Gee JM, Eloranta JJ, Ibbitt JC, Robertson JF, Ellis IO, Williams T, et al. Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol. 2009;217:32–41. - PubMed
-
- Friedrichs N, Jager R, Paggen E, Rudlowski C, Merkelbach-Bruse S, Schorle H, et al. Distinct spatial expression patterns of AP-2alpha and AP-2gamma in non-neoplastic human breast and breast cancer. Mod Pathol. 2005;18:431–438. - PubMed
-
- Woodfield GW, Horan AD, Chen Y, Weigel RJ. TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer Res. 2007;67:8439–8443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

