Insulin stimulates syntaxin4 SNARE complex assembly via a novel regulatory mechanism
- PMID: 24469400
- PMCID: PMC3993566
- DOI: 10.1128/MCB.01203-13
Insulin stimulates syntaxin4 SNARE complex assembly via a novel regulatory mechanism
Abstract
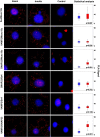
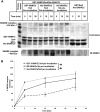
Insulin stimulates glucose transport into fat and muscle cells by increasing the exocytic trafficking rate of the GLUT4 facilitative glucose transporter from intracellular stores to the plasma membrane. Delivery of GLUT4 to the plasma membrane is mediated by formation of functional SNARE complexes containing syntaxin4, SNAP23, and VAMP2. Here we have used an in situ proximity ligation assay to integrate these two observations by demonstrating for the first time that insulin stimulation causes an increase in syntaxin4-containing SNARE complex formation in adipocytes. Furthermore, we demonstrate that insulin brings about this increase in SNARE complex formation by mobilizing a pool of syntaxin4 held in an inactive state under basal conditions. Finally, we have identified phosphorylation of the regulatory protein Munc18c, a direct target of the insulin receptor, as a molecular switch to coordinate this process. Hence, this report provides molecular detail of how the cell alters membrane traffic in response to an external stimulus, in this case, insulin.
Figures


Similar articles
-
The t-SNAREs syntaxin4 and SNAP23 but not v-SNARE VAMP2 are indispensable to tether GLUT4 vesicles at the plasma membrane in adipocyte.Biochem Biophys Res Commun. 2010 Jan 15;391(3):1336-41. doi: 10.1016/j.bbrc.2009.12.045. Epub 2009 Dec 16. Biochem Biophys Res Commun. 2010. PMID: 20006577
-
Molecular dissection of the Munc18c/syntaxin4 interaction: implications for regulation of membrane trafficking.Traffic. 2006 Oct;7(10):1408-19. doi: 10.1111/j.1600-0854.2006.00474.x. Epub 2006 Aug 10. Traffic. 2006. PMID: 16899085
-
Role of SNAP23 in insulin-induced translocation of GLUT4 in 3T3-L1 adipocytes. Mediation of complex formation between syntaxin4 and VAMP2.J Biol Chem. 2000 Mar 17;275(11):8240-7. doi: 10.1074/jbc.275.11.8240. J Biol Chem. 2000. PMID: 10713150
-
Studies of the regulated assembly of SNARE complexes in adipocytes.Biochem Soc Trans. 2014 Oct;42(5):1396-400. doi: 10.1042/BST20140114. Biochem Soc Trans. 2014. PMID: 25233421 Review.
-
SNARE proteins underpin insulin-regulated GLUT4 traffic.Traffic. 2011 Jun;12(6):657-64. doi: 10.1111/j.1600-0854.2011.01163.x. Epub 2011 Feb 8. Traffic. 2011. PMID: 21226814 Review.
Cited by
-
Differential Effects of Munc18s on Multiple Degranulation-Relevant Trans-SNARE Complexes.PLoS One. 2015 Sep 18;10(9):e0138683. doi: 10.1371/journal.pone.0138683. eCollection 2015. PLoS One. 2015. PMID: 26384026 Free PMC article.
-
Evidence for a conserved inhibitory binding mode between the membrane fusion assembly factors Munc18 and syntaxin in animals.J Biol Chem. 2017 Dec 15;292(50):20449-20460. doi: 10.1074/jbc.M117.811182. Epub 2017 Oct 18. J Biol Chem. 2017. PMID: 29046354 Free PMC article.
-
GLUT4 On the move.Biochem J. 2022 Feb 11;479(3):445-462. doi: 10.1042/BCJ20210073. Biochem J. 2022. PMID: 35147164 Free PMC article.
-
Diverse exocytic pathways for mast cell mediators.Biochem Soc Trans. 2018 Apr 17;46(2):235-247. doi: 10.1042/BST20170450. Epub 2018 Feb 22. Biochem Soc Trans. 2018. PMID: 29472369 Free PMC article. Review.
-
An Emerging Role for SNARE Proteins in Dendritic Cell Function.Front Immunol. 2015 Mar 31;6:133. doi: 10.3389/fimmu.2015.00133. eCollection 2015. Front Immunol. 2015. PMID: 25873919 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
