TRF2-tethered TIN2 can mediate telomere protection by TPP1/POT1
- PMID: 24469404
- PMCID: PMC3993560
- DOI: 10.1128/MCB.01052-13
TRF2-tethered TIN2 can mediate telomere protection by TPP1/POT1
Abstract
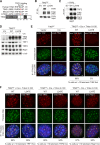
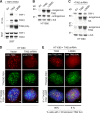
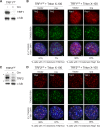
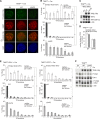
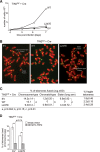
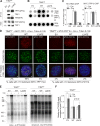
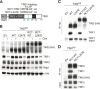
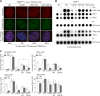
The shelterin protein TIN2 is required for the telomeric accumulation of TPP1/POT1 heterodimers and for the protection of telomeres by the POT1 proteins (POT1a and POT1b in the mouse). TIN2 also binds to TRF1 and TRF2, improving the telomeric localization of TRF2 and its function. Here, we ask whether TIN2 needs to interact with both TRF1 and TRF2 to mediate the telomere protection afforded by TRF2 and POT1a/b. Using a TIN2 allele deficient in TRF1 binding (TIN2-L247E), we demonstrate that TRF1 is required for optimal recruitment of TIN2 to telomeres and document phenotypes associated with the TIN2-L247E allele that are explained by insufficient TIN2 loading onto telomeres. To bypass the requirement for TRF1-dependent recruitment, we fused TIN2-L247E to the TRF2-interacting (RCT) domain of Rap1. The RCT-TIN2-L247E fusion showed improved telomeric localization and was fully functional in terms of chromosome end protection by TRF2, TPP1/POT1a, and TPP1/POT1b. These data indicate that when sufficient TIN2 is loaded onto telomeres, its interaction with TRF1 is not required to mediate the function of TRF2 and the TPP1/POT1 heterodimers. We therefore conclude that shelterin can protect chromosome ends as a TRF2-tethered TIN2/TPP1/POT1 complex that lacks a physical connection to TRF1.
Figures


Similar articles
-
Binding of TPP1 protein to TIN2 protein is required for POT1a,b protein-mediated telomere protection.J Biol Chem. 2014 Aug 29;289(35):24180-7. doi: 10.1074/jbc.M114.592592. Epub 2014 Jul 23. J Biol Chem. 2014. PMID: 25056954 Free PMC article.
-
Human Telomere Repeat Binding Factor TRF1 Replaces TRF2 Bound to Shelterin Core Hub TIN2 when TPP1 Is Absent.J Mol Biol. 2019 Aug 9;431(17):3289-3301. doi: 10.1016/j.jmb.2019.05.038. Epub 2019 May 31. J Mol Biol. 2019. PMID: 31158366
-
Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex.Cell Res. 2017 Dec;27(12):1485-1502. doi: 10.1038/cr.2017.144. Epub 2017 Nov 21. Cell Res. 2017. PMID: 29160297 Free PMC article.
-
Shelterin proteins and cancer.Asian Pac J Cancer Prev. 2015;16(8):3085-90. doi: 10.7314/apjcp.2015.16.8.3085. Asian Pac J Cancer Prev. 2015. PMID: 25921101 Review.
-
Shelterin: the protein complex that shapes and safeguards human telomeres.Genes Dev. 2005 Sep 15;19(18):2100-10. doi: 10.1101/gad.1346005. Genes Dev. 2005. PMID: 16166375 Review.
Cited by
-
The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres.PLoS Genet. 2015 Jul 31;11(7):e1005410. doi: 10.1371/journal.pgen.1005410. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26230315 Free PMC article.
-
HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2.Sci Rep. 2016 Oct 26;6:36045. doi: 10.1038/srep36045. Sci Rep. 2016. PMID: 27782152 Free PMC article.
-
Single-molecule imaging of genome maintenance proteins encountering specific DNA sequences and structures.DNA Repair (Amst). 2023 Aug;128:103528. doi: 10.1016/j.dnarep.2023.103528. Epub 2023 Jun 24. DNA Repair (Amst). 2023. PMID: 37392578 Free PMC article. Review.
-
The Connection Between Cell Fate and Telomere.Adv Exp Med Biol. 2021;1275:71-100. doi: 10.1007/978-3-030-49844-3_3. Adv Exp Med Biol. 2021. PMID: 33539012 Review.
-
Molecular basis and quantitative assessment of TRF1 and TRF2 protein interactions with TIN2 and Apollo peptides.Eur Biophys J. 2017 Mar;46(2):171-187. doi: 10.1007/s00249-016-1157-7. Epub 2016 Jul 22. Eur Biophys J. 2017. PMID: 27450562
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
