Septin assemblies form by diffusion-driven annealing on membranes
- PMID: 24469790
- PMCID: PMC3926015
- DOI: 10.1073/pnas.1314138111
Septin assemblies form by diffusion-driven annealing on membranes
Abstract
Septins assemble into filaments and higher-order structures that act as scaffolds for diverse cell functions including cytokinesis, cell polarity, and membrane remodeling. Despite their conserved role in cell organization, little is known about how septin filaments elongate and are knitted together into higher-order assemblies. Using fluorescence correlation spectroscopy, we determined that cytosolic septins are in small complexes, suggesting that septin filaments are not formed in the cytosol. When the plasma membrane of live cells is monitored by total internal reflection fluorescence microscopy, we see that septin complexes of variable size diffuse in two dimensions. Diffusing septin complexes collide and make end-on associations to form elongated filaments and higher-order structures, an assembly process we call annealing. Septin assembly by annealing can be reconstituted in vitro on supported lipid bilayers with purified septin complexes. Using the reconstitution assay, we show that septin filaments are highly flexible, grow only from free filament ends, and do not exchange subunits in the middle of filaments. This work shows that annealing is a previously unidentified intrinsic property of septins in the presence of membranes and demonstrates that cells exploit this mechanism to build large septin assemblies.
Keywords: biophysics; cytoskeleton.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
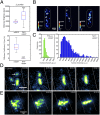
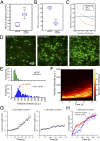
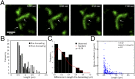

Comment in
-
Cytoskeleton: septins do the horizontal tango.Curr Biol. 2014 Apr 14;24(8):R324-7. doi: 10.1016/j.cub.2014.02.047. Curr Biol. 2014. PMID: 24735857
References
-
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305(5682):393–396. - PubMed
-
- Culotti J, Hartwell LH. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp Cell Res. 1971;67(2):389–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

