Distortion of genealogical properties when the sample is very large
- PMID: 24469801
- PMCID: PMC3926037
- DOI: 10.1073/pnas.1322709111
Distortion of genealogical properties when the sample is very large
Abstract
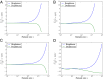
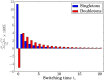
Study sample sizes in human genetics are growing rapidly, and in due course it will become routine to analyze samples with hundreds of thousands, if not millions, of individuals. In addition to posing computational challenges, such large sample sizes call for carefully reexamining the theoretical foundation underlying commonly used analytical tools. Here, we study the accuracy of the coalescent, a central model for studying the ancestry of a sample of individuals. The coalescent arises as a limit of a large class of random mating models, and it is an accurate approximation to the original model provided that the population size is sufficiently larger than the sample size. We develop a method for performing exact computation in the discrete-time Wright-Fisher (DTWF) model and compare several key genealogical quantities of interest with the coalescent predictions. For recently inferred demographic scenarios, we find that there are a significant number of multiple- and simultaneous-merger events under the DTWF model, which are absent in the coalescent by construction. Furthermore, for large sample sizes, there are noticeable differences in the expected number of rare variants between the coalescent and the DTWF model. To balance the trade-off between accuracy and computational efficiency, we propose a hybrid algorithm that uses the DTWF model for the recent past and the coalescent for the more distant past. Our results demonstrate that the hybrid method with only a handful of generations of the DTWF model leads to a frequency spectrum that is quite close to the prediction of the full DTWF model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

