Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal
- PMID: 24469802
- PMCID: PMC3926038
- DOI: 10.1073/pnas.1318934111
Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal
Abstract
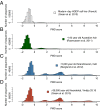
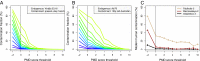
One of the main impediments for obtaining DNA sequences from ancient human skeletons is the presence of contaminating modern human DNA molecules in many fossil samples and laboratory reagents. However, DNA fragments isolated from ancient specimens show a characteristic DNA damage pattern caused by miscoding lesions that differs from present day DNA sequences. Here, we develop a framework for evaluating the likelihood of a sequence originating from a model with postmortem degradation-summarized in a postmortem degradation score-which allows the identification of DNA fragments that are unlikely to originate from present day sources. We apply this approach to a contaminated Neandertal specimen from Okladnikov Cave in Siberia to isolate its endogenous DNA from modern human contaminants and show that the reconstructed mitochondrial genome sequence is more closely related to the variation of Western Neandertals than what was discernible from previous analyses. Our method opens up the potential for genomic analysis of contaminated fossil material.
Keywords: human evolution; paleogenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

