On the dephasing of genetic oscillators
- PMID: 24469814
- PMCID: PMC3926054
- DOI: 10.1073/pnas.1323433111
On the dephasing of genetic oscillators
Abstract
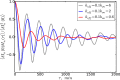
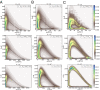
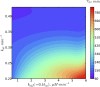
The digital nature of genes combined with the associated low copy numbers of proteins regulating them is a significant source of stochasticity, which affects the phase of biochemical oscillations. We show that unlike ordinary chemical oscillators, the dichotomic molecular noise of gene state switching in gene oscillators affects the stochastic dephasing in a way that may not always be captured by phenomenological limit cycle-based models. Through simulations of a realistic model of the NFκB/IκB network, we also illustrate the dephasing phenomena that are important for reconciling single-cell and population-based experiments on gene oscillators.
Keywords: gene networks; master equation; stochastic simulations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


 , and the ratio of oscillation frequency
, and the ratio of oscillation frequency  to decay rate γ. Different points correspond to different eigenvalues. Slow modes are grouped around
to decay rate γ. Different points correspond to different eigenvalues. Slow modes are grouped around  . Dominant rotational modes have a high
. Dominant rotational modes have a high  ratio.
ratio.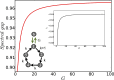

References
-
- Winfree A. The Geometry of Biological Time. New York: Springer; 2001.
-
- Morelli LG, Jülicher F. Precision of genetic oscillators and clocks. Phys Rev Lett. 2007;98(22):228101. - PubMed
-
- Goldbeter A. Biochemical Oscillations and Cellular Rhythms. Cambridge, UK: Cambridge Univ Press; 1997.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

