Identifying and mapping cell-type-specific chromatin programming of gene expression
- PMID: 24469817
- PMCID: PMC3926062
- DOI: 10.1073/pnas.1312523111
Identifying and mapping cell-type-specific chromatin programming of gene expression
Abstract
A problem of substantial interest is to systematically map variation in chromatin structure to gene-expression regulation across conditions, environments, or differentiated cell types. We developed and applied a quantitative framework for determining the existence, strength, and type of relationship between high-resolution chromatin structure in terms of DNaseI hypersensitivity and genome-wide gene-expression levels in 20 diverse human cell types. We show that ∼25% of genes show cell-type-specific expression explained by alterations in chromatin structure. We find that distal regions of chromatin structure (e.g., ±200 kb) capture more genes with this relationship than local regions (e.g., ±2.5 kb), yet the local regions show a more pronounced effect. By exploiting variation across cell types, we were capable of pinpointing the most likely hypersensitive sites related to cell-type-specific expression, which we show have a range of contextual uses. This quantitative framework is likely applicable to other settings aimed at relating continuous genomic measurements to gene-expression variation.
Keywords: association; computational biology; encode; epigenetics; gene regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

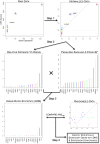
 . The maximal statistic
. The maximal statistic  is calculated for each gene and the corresponding cell type recorded, in this case the TH1 cell line. (Step 4) A randomization method is performed to generate null data, upon which null
is calculated for each gene and the corresponding cell type recorded, in this case the TH1 cell line. (Step 4) A randomization method is performed to generate null data, upon which null  are calculated. These are compared with the observed
are calculated. These are compared with the observed  values to calculate the statistical significance of each gene.
values to calculate the statistical significance of each gene.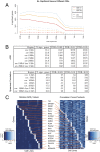
 (
( , as estimated in ref. 13). Columns 3–5 show the number of statistically significant genes at various FDR cutoffs. Although the 2.5-kb window shows more significant genes at the stringent FDR cutoffs, indicating a larger effect size, the overall percentage of genes showing a relationship is notably lower than the more distal DHS volumes. Compared with Spearman correlation, ARS is more powerful at detecting these associations (see
, as estimated in ref. 13). Columns 3–5 show the number of statistically significant genes at various FDR cutoffs. Although the 2.5-kb window shows more significant genes at the stringent FDR cutoffs, indicating a larger effect size, the overall percentage of genes showing a relationship is notably lower than the more distal DHS volumes. Compared with Spearman correlation, ARS is more powerful at detecting these associations (see 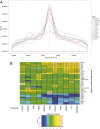
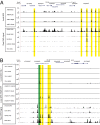
References
-
- Satterlee JS, Schübeler D, Ng HH. Tackling the epigenome: Challenges and opportunities for collaboration. Nat Biotechnol. 2010;28(10):1039–1044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

