Stability-activity tradeoffs constrain the adaptive evolution of RubisCO
- PMID: 24469821
- PMCID: PMC3926066
- DOI: 10.1073/pnas.1310811111
Stability-activity tradeoffs constrain the adaptive evolution of RubisCO
Abstract
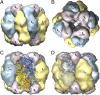
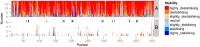
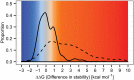
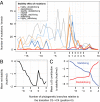
A well-known case of evolutionary adaptation is that of ribulose-1,5-bisphosphate carboxylase (RubisCO), the enzyme responsible for fixation of CO2 during photosynthesis. Although the majority of plants use the ancestral C3 photosynthetic pathway, many flowering plants have evolved a derived pathway named C4 photosynthesis. The latter concentrates CO2, and C4 RubisCOs consequently have lower specificity for, and faster turnover of, CO2. The C4 forms result from convergent evolution in multiple clades, with substitutions at a small number of sites under positive selection. To understand the physical constraints on these evolutionary changes, we reconstructed in silico ancestral sequences and 3D structures of RubisCO from a large group of related C3 and C4 species. We were able to precisely track their past evolutionary trajectories, identify mutations on each branch of the phylogeny, and evaluate their stability effect. We show that RubisCO evolution has been constrained by stability-activity tradeoffs similar in character to those previously identified in laboratory-based experiments. The C4 properties require a subset of several ancestral destabilizing mutations, which from their location in the structure are inferred to mainly be involved in enhancing conformational flexibility of the open-closed transition in the catalytic cycle. These mutations are near, but not in, the active site or at intersubunit interfaces. The C3 to C4 transition is preceded by a sustained period in which stability of the enzyme is increased, creating the capacity to accept the functionally necessary destabilizing mutations, and is immediately followed by compensatory mutations that restore global stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme.Mol Biol Evol. 2011 Apr;28(4):1491-503. doi: 10.1093/molbev/msq335. Epub 2010 Dec 16. Mol Biol Evol. 2011. PMID: 21172830
-
Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis.Mol Biol Evol. 2008 Nov;25(11):2361-8. doi: 10.1093/molbev/msn178. Epub 2008 Aug 11. Mol Biol Evol. 2008. PMID: 18695049
-
Kranz and single-cell forms of C4 plants in the subfamily Suaedoideae show kinetic C4 convergence for PEPC and Rubisco with divergent amino acid substitutions.J Exp Bot. 2015 Dec;66(22):7347-58. doi: 10.1093/jxb/erv431. Epub 2015 Sep 28. J Exp Bot. 2015. PMID: 26417023 Free PMC article.
-
Deconstructing Kranz anatomy to understand C4 evolution.J Exp Bot. 2014 Jul;65(13):3357-69. doi: 10.1093/jxb/eru186. Epub 2014 May 5. J Exp Bot. 2014. PMID: 24799561 Review.
-
Prospects for improving CO2 fixation in C3-crops through understanding C4-Rubisco biogenesis and catalytic diversity.Curr Opin Plant Biol. 2016 Jun;31:135-42. doi: 10.1016/j.pbi.2016.04.002. Epub 2016 Apr 27. Curr Opin Plant Biol. 2016. PMID: 27131319 Review.
Cited by
-
Bacterial Hsp90 predominantly buffers but does not potentiate the phenotypic effects of deleterious mutations during fluorescent protein evolution.Genetics. 2022 Nov 30;222(4):iyac154. doi: 10.1093/genetics/iyac154. Genetics. 2022. PMID: 36227141 Free PMC article.
-
Exceptionally high rates of positive selection on the rbcL gene in the genus Ilex (Aquifoliaceae).BMC Evol Biol. 2019 Oct 21;19(1):192. doi: 10.1186/s12862-019-1521-1. BMC Evol Biol. 2019. PMID: 31638910 Free PMC article.
-
Response: Commentary: Directions for Optimization of Photosynthetic Carbon Fixation: RuBisCO's Efficiency May Not Be So Constrained After All.Front Plant Sci. 2019 Nov 22;10:1426. doi: 10.3389/fpls.2019.01426. eCollection 2019. Front Plant Sci. 2019. PMID: 31824523 Free PMC article. No abstract available.
-
Adaptive HIV-1 evolutionary trajectories are constrained by protein stability.Virus Evol. 2017 Aug 1;3(2):vex019. doi: 10.1093/ve/vex019. eCollection 2017 Jul. Virus Evol. 2017. PMID: 28852572 Free PMC article.
-
Ancestral Reconstruction and the Evolution of Protein Energy Landscapes.Annu Rev Biophys. 2024 Jul;53(1):127-146. doi: 10.1146/annurev-biophys-030722-125440. Epub 2024 Jun 28. Annu Rev Biophys. 2024. PMID: 38134334 Free PMC article. Review.
References
-
- DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6(9):678–687. - PubMed
-
- Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins. 2002;46(1):105–109. - PubMed
-
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19(5):596–604. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

