Transcriptional control of early T and B cell developmental choices
- PMID: 24471430
- PMCID: PMC3994230
- DOI: 10.1146/annurev-immunol-032712-100024
Transcriptional control of early T and B cell developmental choices
Abstract
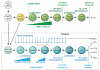
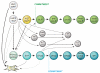
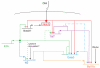
T and B cells share a common somatic gene rearrangement mechanism for assembling the genes that code for their antigen receptors; they also have developmental pathways with many parallels. Shared usage of basic helix-loop-helix E proteins as transcriptional drivers underlies these common features. However, the transcription factor networks in which these E proteins are embedded are different both in membership and in architecture for T and B cell gene regulatory programs. These differences permit lineage commitment decisions to be made in different hierarchical orders. Furthermore, in contrast to B cell gene networks, the T cell gene network architecture for effector differentiation is sufficiently modular so that E protein inputs can be removed. Complete T cell-like effector differentiation can proceed without T cell receptor rearrangement or selection when E proteins are neutralized, yielding natural killer and other innate lymphoid cells.
Figures

References
-
- Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, Venkatesh B, Yu JK, Kaltenbach SL, Holland ND, Diekhoff D, Happe C, Schorpp M, Boehm T. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–97. - PubMed
-
- Zandi S, Bryder D, Sigvardsson M. Load and lock: the molecular mechanisms of B-lymphocyte commitment. Immunol Rev. 2010;238:47–62. - PubMed
-
- Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr.Opin.Immunol. 2010;22:161–7. - PubMed
-
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat.Immunol. 2007;8:463–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

