Investigation of bacterial nucleotide excision repair using single-molecule techniques
- PMID: 24472181
- PMCID: PMC5053424
- DOI: 10.1016/j.dnarep.2013.10.012
Investigation of bacterial nucleotide excision repair using single-molecule techniques
Abstract
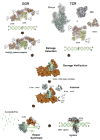
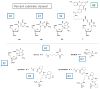
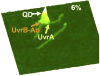
Despite three decades of biochemical and structural analysis of the prokaryotic nucleotide excision repair (NER) system, many intriguing questions remain with regard to how the UvrA, UvrB, and UvrC proteins detect, verify and remove a wide range of DNA lesions. Single-molecule techniques have begun to allow more detailed understanding of the kinetics and action mechanism of this complex process. This article reviews how atomic force microscopy and fluorescence microscopy have captured new glimpses of how these proteins work together to mediate NER.
Keywords: Bacterial nucleotide excision repair; Single molecule; UvrA; UvrB; UvrC; UvrD.
Copyright © 2014 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
statement We have no conflict of interest.
Figures



Similar articles
-
A combined structural and biochemical approach reveals translocation and stalling of UvrB on the DNA lesion as a mechanism of damage verification in bacterial nucleotide excision repair.DNA Repair (Amst). 2020 Jan;85:102746. doi: 10.1016/j.dnarep.2019.102746. Epub 2019 Nov 6. DNA Repair (Amst). 2020. PMID: 31739207 Free PMC article.
-
Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair.EMBO J. 2004 Jul 7;23(13):2498-509. doi: 10.1038/sj.emboj.7600263. Epub 2004 Jun 10. EMBO J. 2004. PMID: 15192705 Free PMC article.
-
Dynamics of the UvrABC nucleotide excision repair proteins analyzed by fluorescence resonance energy transfer.Biochemistry. 2007 Aug 7;46(31):9080-8. doi: 10.1021/bi7002235. Epub 2007 Jul 14. Biochemistry. 2007. PMID: 17630776
-
The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch.Mutat Res. 2000 Aug 30;460(3-4):277-300. doi: 10.1016/s0921-8777(00)00032-x. Mutat Res. 2000. PMID: 10946234 Review.
-
Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair.Res Microbiol. 2001 Apr-May;152(3-4):401-9. doi: 10.1016/s0923-2508(01)01211-6. Res Microbiol. 2001. PMID: 11421287 Review.
Cited by
-
Probing the mechanism of nick searching by LIG1 at the single-molecule level.Nucleic Acids Res. 2024 Nov 11;52(20):12604-12615. doi: 10.1093/nar/gkae865. Nucleic Acids Res. 2024. PMID: 39404052 Free PMC article.
-
Insight into Single-Molecule Imaging Techniques for the Study of Prokaryotic Genome Maintenance.Chem Biomed Imaging. 2024 Jun 18;2(9):595-614. doi: 10.1021/cbmi.4c00037. eCollection 2024 Sep 23. Chem Biomed Imaging. 2024. PMID: 39328428 Free PMC article. Review.
-
Bacterial DNA excision repair pathways.Nat Rev Microbiol. 2022 Aug;20(8):465-477. doi: 10.1038/s41579-022-00694-0. Epub 2022 Feb 24. Nat Rev Microbiol. 2022. PMID: 35210609 Free PMC article. Review.
-
Bacterial phenotypic heterogeneity in DNA repair and mutagenesis.Biochem Soc Trans. 2020 Apr 29;48(2):451-462. doi: 10.1042/BST20190364. Biochem Soc Trans. 2020. PMID: 32196548 Free PMC article. Review.
-
Guidelines for DNA recombination and repair studies: Mechanistic assays of DNA repair processes.Microb Cell. 2019 Jan 7;6(1):65-101. doi: 10.15698/mic2019.01.665. Microb Cell. 2019. PMID: 30652106 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

