A tool for design of primers for microRNA-specific quantitative RT-qPCR
- PMID: 24472427
- PMCID: PMC3922658
- DOI: 10.1186/1471-2105-15-29
A tool for design of primers for microRNA-specific quantitative RT-qPCR
Abstract
Background: MicroRNAs are small but biologically important RNA molecules. Although different methods can be used for quantification of microRNAs, quantitative PCR is regarded as the reference that is used to validate other methods. Several commercial qPCR assays are available but they often come at a high price and the sequences of the primers are not disclosed. An alternative to commercial assays is to manually design primers but this work is tedious and, hence, not practical for the design of primers for a larger number of targets.
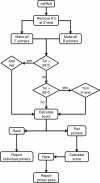
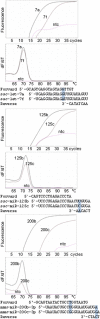
Results: I have developed the software miRprimer for automatic design of primers for the method miR-specific RT-qPCR, which is one of the best performing microRNA qPCR methods available. The algorithm is based on an implementation of the previously published rules for manual design of miR-specific primers with the additional feature of evaluating the propensity of formation of secondary structures and primer dimers. Testing of the primers showed that 76 out of 79 primers (96%) worked for quantification of microRNAs by miR-specific RT-qPCR of mammalian RNA samples. This success rate corresponds to the success rate of manual primer design. Furthermore, primers designed by this method have been distributed to several labs and used successfully in published studies.
Conclusions: The software miRprimer is an automatic and easy method for design of functional primers for miR-specific RT-qPCR. The application is available as stand-alone software that will work on the MS Windows platform and in a developer version written in the Ruby programming language.
Figures


References
-
- Kolbert CP, Feddersen RM, Rakhshan F, Grill DE, Simon G, Middha S, Jang JS, Simon V, Schultz DA, Zschunke M, Lingle W, Carr JM, Thompson EA, Oberg AL, Eckloff BW, Wieben ED, Li P, Yang P, Jen J. Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS ONE. 2013;8:e52517. doi: 10.1371/journal.pone.0052517. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

