Two- and three-dimensional rings in drugs
- PMID: 24472495
- PMCID: PMC4233953
- DOI: 10.1111/cbdd.12260
Two- and three-dimensional rings in drugs
Abstract
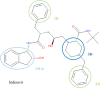
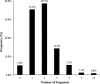
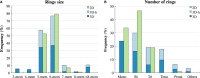
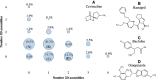
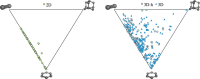
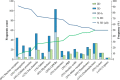
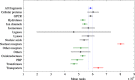
Using small, flat aromatic rings as components of fragments or molecules is a common practice in fragment-based drug discovery and lead optimization. With an increasing focus on the exploration of novel biological and chemical space, and their improved synthetic accessibility, 3D fragments are attracting increasing interest. This study presents a detailed analysis of 3D and 2D ring fragments in marketed drugs. Several measures of properties were used, such as the type of ring assemblies and molecular shapes. The study also took into account the relationship between protein classes targeted by each ring fragment, providing target-specific information. The analysis shows the high structural and shape diversity of 3D ring systems and their importance in bioactive compounds. Major differences in 2D and 3D fragments are apparent in ligands that bind to the major drug targets such as GPCRs, ion channels, and enzymes.
Keywords: drug; fragment; lead optimization; ring.
© 2013 The Authors Chemical Biology & Drug Design Published by John Wiley & Sons Ltd.
Figures

References
-
- Murray CW, Verdonk ML, Rees DC. Experiences in fragment-based drug discovery. Trends Pharmacol Sci. 2012;33:224–232. - PubMed
-
- Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. - PubMed
-
- Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov Today. 2003;8:876–877. - PubMed
-
- Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem. 2008;51:3661–3680. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

