Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems
- PMID: 24473129
- PMCID: PMC3903277
- DOI: 10.1128/mBio.00928-13
Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems
Abstract
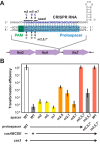
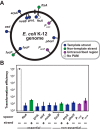
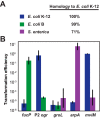
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems in bacteria and archaea employ CRISPR RNAs to specifically recognize the complementary DNA of foreign invaders, leading to sequence-specific cleavage or degradation of the target DNA. Recent work has shown that the accidental or intentional targeting of the bacterial genome is cytotoxic and can lead to cell death. Here, we have demonstrated that genome targeting with CRISPR-Cas systems can be employed for the sequence-specific and titratable removal of individual bacterial strains and species. Using the type I-E CRISPR-Cas system in Escherichia coli as a model, we found that this effect could be elicited using native or imported systems and was similarly potent regardless of the genomic location, strand, or transcriptional activity of the target sequence. Furthermore, the specificity of targeting with CRISPR RNAs could readily distinguish between even highly similar strains in pure or mixed cultures. Finally, varying the collection of delivered CRISPR RNAs could quantitatively control the relative number of individual strains within a mixed culture. Critically, the observed selectivity and programmability of bacterial removal would be virtually impossible with traditional antibiotics, bacteriophages, selectable markers, or tailored growth conditions. Once delivery challenges are addressed, we envision that this approach could offer a novel means to quantitatively control the composition of environmental and industrial microbial consortia and may open new avenues for the development of "smart" antibiotics that circumvent multidrug resistance and differentiate between pathogenic and beneficial microorganisms.
Importance: Controlling the composition of microbial populations is a critical aspect in medicine, biotechnology, and environmental cycles. While different antimicrobial strategies, such as antibiotics, antimicrobial peptides, and lytic bacteriophages, offer partial solutions, what remains elusive is a generalized and programmable strategy that can distinguish between even closely related microorganisms and that allows for fine control over the composition of a microbial population. This study demonstrates that RNA-directed immune systems in bacteria and archaea called CRISPR-Cas systems can provide such a strategy. These systems can be employed to selectively and quantitatively remove individual bacterial strains based purely on sequence information, creating opportunities in the treatment of multidrug-resistant infections, the control of industrial fermentations, and the study of microbial consortia.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
