Genomic analysis of thermophilic Bacillus coagulans strains: efficient producers for platform bio-chemicals
- PMID: 24473268
- PMCID: PMC3905273
- DOI: 10.1038/srep03926
Genomic analysis of thermophilic Bacillus coagulans strains: efficient producers for platform bio-chemicals
Abstract
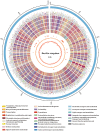
Microbial strains with high substrate efficiency and excellent environmental tolerance are urgently needed for the production of platform bio-chemicals. Bacillus coagulans has these merits; however, little genetic information is available about this species. Here, we determined the genome sequences of five B. coagulans strains, and used a comparative genomic approach to reconstruct the central carbon metabolism of this species to explain their fermentation features. A novel xylose isomerase in the xylose utilization pathway was identified in these strains. Based on a genome-wide positive selection scan, the selection pressure on amino acid metabolism may have played a significant role in the thermal adaptation. We also researched the immune systems of B. coagulans strains, which provide them with acquired resistance to phages and mobile genetic elements. Our genomic analysis provides comprehensive insights into the genetic characteristics of B. coagulans and paves the way for improving and extending the uses of this species.
Figures
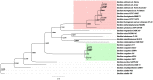

 ) are from B. coagulans strains and are homologs of xylA in B. subtilis. The genes marked with squares (
) are from B. coagulans strains and are homologs of xylA in B. subtilis. The genes marked with squares ( ) are from B. coagulans strains, and are the novel xylose isomerases discovered in this study. (D) Maximum likelihood tree of xylB genes. The genes marked with filled circle (
) are from B. coagulans strains, and are the novel xylose isomerases discovered in this study. (D) Maximum likelihood tree of xylB genes. The genes marked with filled circle ( ) are from B. coagulans strains. The accession numbers of these genes downloaded from the NCBI database are shown in the parentheses. A scale bar for the genetic distance is shown at the bottom.
) are from B. coagulans strains. The accession numbers of these genes downloaded from the NCBI database are shown in the parentheses. A scale bar for the genetic distance is shown at the bottom.

Similar articles
-
Expanded genome and proteome reallocation in a novel, robust Bacillus coagulans strain capable of utilizing pentose and hexose sugars.mSystems. 2024 Nov 19;9(11):e0095224. doi: 10.1128/msystems.00952-24. Epub 2024 Oct 8. mSystems. 2024. PMID: 39377583 Free PMC article.
-
Genome sequence of the thermophilic strain Bacillus coagulans 2-6, an efficient producer of high-optical-purity L-lactic acid.J Bacteriol. 2011 Sep;193(17):4563-4. doi: 10.1128/JB.05378-11. Epub 2011 Jun 24. J Bacteriol. 2011. PMID: 21705584 Free PMC article.
-
Genome sequence of the thermophilic strain Bacillus coagulans XZL4, an efficient pentose-utilizing producer of chemicals.J Bacteriol. 2011 Nov;193(22):6398-9. doi: 10.1128/JB.06157-11. J Bacteriol. 2011. PMID: 22038963 Free PMC article.
-
The Bacillus cereus group: novel aspects of population structure and genome dynamics.J Appl Microbiol. 2006 Sep;101(3):579-93. doi: 10.1111/j.1365-2672.2006.03087.x. J Appl Microbiol. 2006. PMID: 16907808 Review.
-
Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol.Appl Microbiol Biotechnol. 2015 Jan;99(2):535-51. doi: 10.1007/s00253-014-6224-3. Epub 2014 Nov 28. Appl Microbiol Biotechnol. 2015. PMID: 25431011 Review.
Cited by
-
A cold shock protein promotes high-temperature microbial growth through binding to diverse RNA species.Cell Discov. 2021 Mar 16;7(1):15. doi: 10.1038/s41421-021-00246-5. Cell Discov. 2021. PMID: 33727528 Free PMC article.
-
Expanded genome and proteome reallocation in a novel, robust Bacillus coagulans strain capable of utilizing pentose and hexose sugars.mSystems. 2024 Nov 19;9(11):e0095224. doi: 10.1128/msystems.00952-24. Epub 2024 Oct 8. mSystems. 2024. PMID: 39377583 Free PMC article.
-
Engineered microbial platform confers resistance against heavy metals via phosphomelanin biosynthesis.Nat Commun. 2025 May 24;16(1):4836. doi: 10.1038/s41467-025-60117-5. Nat Commun. 2025. PMID: 40413165 Free PMC article.
-
Progress of research and application of Heyndrickxia coagulans (Bacillus coagulans) as probiotic bacteria.Front Cell Infect Microbiol. 2024 May 28;14:1415790. doi: 10.3389/fcimb.2024.1415790. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38863834 Free PMC article. Review.
-
Genomic analysis of a ginger pathogen Bacillus pumilus providing the understanding to the pathogenesis and the novel control strategy.Sci Rep. 2015 May 19;5:10259. doi: 10.1038/srep10259. Sci Rep. 2015. PMID: 25989507 Free PMC article.
References
-
- Lorenz P. & Zinke H. White biotechnology: differences in US and EU approaches? Trends Biotechnol. 23, 570–574 (2005). - PubMed
-
- Gao C., Ma C. & Xu P. Biotechnological routes based on lactic acid production from biomass. Biotechnol. Adv. 29, 930–939 (2011). - PubMed
-
- Teusink B. & Smid E. Modelling strategies for the industrial exploitation of lactic acid bacteria. Nat. Rev. Microbiol. 4, 46–56 (2006). - PubMed
-
- Zhao B. et al. Repeated open fermentative production of optically pure l-lactic acid using a thermophilic Bacillus sp. strain. Bioresour. Technol. 101, 6494–6498 (2010). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

