Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease
- PMID: 24474917
- PMCID: PMC3893567
- DOI: 10.3389/fninf.2013.00050
Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease
Abstract
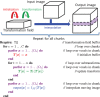
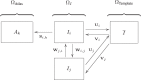
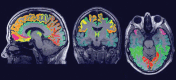
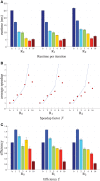
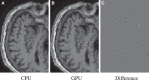
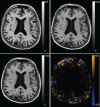
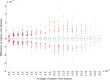
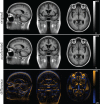
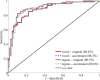
Nonrigid image registration is an important, but time-consuming task in medical image analysis. In typical neuroimaging studies, multiple image registrations are performed, i.e., for atlas-based segmentation or template construction. Faster image registration routines would therefore be beneficial. In this paper we explore acceleration of the image registration package elastix by a combination of several techniques: (i) parallelization on the CPU, to speed up the cost function derivative calculation; (ii) parallelization on the GPU building on and extending the OpenCL framework from ITKv4, to speed up the Gaussian pyramid computation and the image resampling step; (iii) exploitation of certain properties of the B-spline transformation model; (iv) further software optimizations. The accelerated registration tool is employed in a study on diagnostic classification of Alzheimer's disease and cognitively normal controls based on T1-weighted MRI. We selected 299 participants from the publicly available Alzheimer's Disease Neuroimaging Initiative database. Classification is performed with a support vector machine based on gray matter volumes as a marker for atrophy. We evaluated two types of strategies (voxel-wise and region-wise) that heavily rely on nonrigid image registration. Parallelization and optimization resulted in an acceleration factor of 4-5x on an 8-core machine. Using OpenCL a speedup factor of 2 was realized for computation of the Gaussian pyramids, and 15-60 for the resampling step, for larger images. The voxel-wise and the region-wise classification methods had an area under the receiver operator characteristic curve of 88 and 90%, respectively, both for standard and accelerated registration. We conclude that the image registration package elastix was substantially accelerated, with nearly identical results to the non-optimized version. The new functionality will become available in the next release of elastix as open source under the BSD license.
Keywords: Alzheimer's disease; OpenCL; acceleration; elastix; image registration; parallelization.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

