Transcriptomic meta-analysis of multiple sclerosis and its experimental models
- PMID: 24475162
- PMCID: PMC3903571
- DOI: 10.1371/journal.pone.0086643
Transcriptomic meta-analysis of multiple sclerosis and its experimental models
Abstract
Background: Multiple microarray analyses of multiple sclerosis (MS) and its experimental models have been published in the last years.
Objective: Meta-analyses integrate the information from multiple studies and are suggested to be a powerful approach in detecting highly relevant and commonly affected pathways.
Data sources: ArrayExpress, Gene Expression Omnibus and PubMed databases were screened for microarray gene expression profiling studies of MS and its experimental animal models.
Study eligibility criteria: Studies comparing central nervous system (CNS) samples of diseased versus healthy individuals with n >1 per group and publically available raw data were selected.
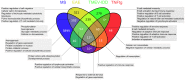
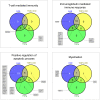
Material and methods: Included conditions for re-analysis of differentially expressed genes (DEGs) were MS, myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis (EAE) in rats, proteolipid protein-induced EAE in mice, Theiler's murine encephalomyelitis virus-induced demyelinating disease (TMEV-IDD), and a transgenic tumor necrosis factor-overexpressing mouse model (TNFtg). Since solely a single MS raw data set fulfilled the inclusion criteria, a merged list containing the DEGs from two MS-studies was additionally included. Cross-study analysis was performed employing list comparisons of DEGs and alternatively Gene Set Enrichment Analysis (GSEA).
Results: The intersection of DEGs in MS, EAE, TMEV-IDD, and TNFtg contained 12 genes related to macrophage functions. The intersection of EAE, TMEV-IDD and TNFtg comprised 40 DEGs, functionally related to positive regulation of immune response. Over and above, GSEA identified substantially more differentially regulated pathways including coagulation and JAK/STAT-signaling.
Conclusion: A meta-analysis based on a simple comparison of DEGs is over-conservative. In contrast, the more experimental GSEA approach identified both, a priori anticipated as well as promising new candidate pathways.
Conflict of interest statement
Figures


References
-
- Comabella M, Martin R (2007) Genomics in multiple sclerosis - Current state and future directions. J Neuroimmunol 187: 1–8. - PubMed
-
- Lindberg RLP, De Groot CJA, Certa U, Ravid R, Hoffmann F, et al. (2004) Multiple sclerosis as a generalized CNS disease - comparative microarray analysis of normal appearing white matter and lesions in secondary progressive MS. J Neuroimmunol 152: 154–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

