Mycobacterium tuberculosis subverts the TLR-2-MyD88 pathway to facilitate its translocation into the cytosol
- PMID: 24475192
- PMCID: PMC3903598
- DOI: 10.1371/journal.pone.0086886
Mycobacterium tuberculosis subverts the TLR-2-MyD88 pathway to facilitate its translocation into the cytosol
Abstract
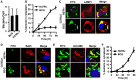
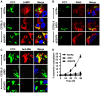
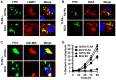
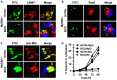
Mycobacterium tuberculosis (M.tb) has evolved mechanisms to evade its destruction in phagolysosomes, where it successfully survives and replicates within phagocytes. Recent studies have shown that virulent strains of M.tb can translocate from the phagosome into the cytosol of dendritic cells (DC). The molecular mechanisms by which virulent M.tb strains can escape the phagosome remain unknown. Here we show that the virulent M.tb strain H37Rv, but not the vaccine strain Bacille Calmette-Guérin (BCG), escapes from the phagolysosome and enters the cytosol by interfering with the TLR-2-MyD88 signaling pathway. Using H37Rv mutants, we further demonstrate that the region of difference-1 (RD-1) locus and ESAT-6, a gene within the RD-1 locus, play an important role in the capacity of M.tb to migrate from the phagosome to the cytosol of macrophages. H37Rv, BCG, H37RvΔRD1, and H37RvΔESAT6 were able to translocate to the cytosol in macrophages derived from TLR-2- and MyD88-deficient animals, whereas only virulent H37Rv was able to enter the cytosol in macrophages from wild type mice. Therefore, signaling through the TLR-2-MyD88 pathway in macrophages plays an important role in confining M.tb within phagolysomes. Virulent strains of M.tb have evolved mechanisms to subvert this pathway, thus facilitating their translocation to the cytosol and to escape the toxic microenvironment of the phagosome or phagolysosome.
Conflict of interest statement
Figures
Similar articles
-
Early secreted antigen ESAT-6 of Mycobacterium tuberculosis promotes protective T helper 17 cell responses in a toll-like receptor-2-dependent manner.PLoS Pathog. 2011 Nov;7(11):e1002378. doi: 10.1371/journal.ppat.1002378. Epub 2011 Nov 10. PLoS Pathog. 2011. PMID: 22102818 Free PMC article.
-
[Frontier of mycobacterium research--host vs. mycobacterium].Kekkaku. 2005 Sep;80(9):613-29. Kekkaku. 2005. PMID: 16245793 Japanese.
-
Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages.Infect Immun. 1993 Jul;61(7):2763-73. doi: 10.1128/iai.61.7.2763-2773.1993. Infect Immun. 1993. PMID: 8514378 Free PMC article.
-
MyDths and un-TOLLed truths: sensor, instructive and effector immunity to tuberculosis.Immunol Lett. 2008 Feb 15;116(1):15-23. doi: 10.1016/j.imlet.2007.11.015. Epub 2007 Dec 26. Immunol Lett. 2008. PMID: 18191460 Review.
-
Natural and trained innate immunity against Mycobacterium tuberculosis.Immunobiology. 2020 May;225(3):151951. doi: 10.1016/j.imbio.2020.151951. Epub 2020 Apr 27. Immunobiology. 2020. PMID: 32423788 Review.
Cited by
-
Host-directed therapy to combat mycobacterial infections.Immunol Rev. 2021 May;301(1):62-83. doi: 10.1111/imr.12951. Epub 2021 Feb 9. Immunol Rev. 2021. PMID: 33565103 Free PMC article. Review.
-
Vitamin D modulates human macrophage response to Mycobacterium tuberculosis DNA.Tuberculosis (Edinb). 2019 May;116S:S131-S137. doi: 10.1016/j.tube.2019.04.021. Epub 2019 May 3. Tuberculosis (Edinb). 2019. PMID: 31085128 Free PMC article.
-
Dynamic post-translational modification profiling of Mycobacterium tuberculosis-infected primary macrophages.Elife. 2020 Jan 17;9:e51461. doi: 10.7554/eLife.51461. Elife. 2020. PMID: 31951200 Free PMC article.
-
The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections.Eur J Clin Microbiol Infect Dis. 2015 May;34(5):963-974. doi: 10.1007/s10096-014-2309-2. Epub 2015 Jan 13. Eur J Clin Microbiol Infect Dis. 2015. PMID: 25579795 Free PMC article.
-
Mcl-1 signals pathway inhibitors in mouse peritoneal macrophage apoptosis infected with the Xinjiang strain of M. tuberculosis.Int J Clin Exp Pathol. 2017 Dec 1;10(12):11952-11967. eCollection 2017. Int J Clin Exp Pathol. 2017. PMID: 31966560 Free PMC article.
References
-
- Vergne I, Chua J, Singh SB, Deretic V (2004) Cell biology of mycobacterium tuberculosis phagosome. Annu Rev Cell Dev Biol 20: 367–394. - PubMed
-
- Rohde K, Yates RM, Purdy GE, Russell DG (2007) Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev 219: 37–54. - PubMed
-
- Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, et al. (1994) Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263: 678–681. - PubMed
-
- Sinai AP, Joiner KA (1997) Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu Rev Microbiol 51: 415–462. - PubMed
-
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, et al. (2007) M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129: 1287–1298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

