Continuing analysis of microRNA origins: Formation from transposable element insertions and noncoding RNA mutations
- PMID: 24475369
- PMCID: PMC3891635
- DOI: 10.4161/mge.27755
Continuing analysis of microRNA origins: Formation from transposable element insertions and noncoding RNA mutations
Abstract
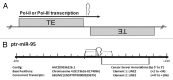
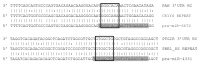
MicroRNAs (miRs) are small noncoding RNAs that typically act as regulators of gene expression by base pairing with the 3' UTR of messenger RNAs (mRNAs) and either repressing their translation or initiating degradation. As of this writing over 24,500 distinct miRs have been identified, but the functions of the vast majority of these remain undescribed. This paper represents a summary of our in depth analysis of the genomic origins of miR loci, detailing the formation of 1,213 of the 7,321 recently identified miRs and thereby bringing the total number of miR loci with defined molecular origin to 3,605. Interestingly, our analyses also identify evidence for a second, novel mechanism of miR locus generation through describing the formation of 273 miR loci from mutations to other forms of noncoding RNAs. Importantly, several independent investigations of the genomic origins of miR loci have now supported the hypothesis that miR hairpins are formed by the adjacent genomic insertion of two complementary transposable elements (TEs) into opposing strands. While our results agree that subsequent transcription over such TE interfaces leads to the formation of the majority of functional miR loci, we now also find evidence suggesting that a subset of miR loci were actually formed by an alternative mechanism-point mutations in other structurally complex, noncoding RNAs (e.g., tRNAs and snoRNAs).
Keywords: LINE; SINE; miR; miRNA; microRNA; noncoding RNA; repetitive; retrotransposon; transposable; transposon.
Figures



References
-
- Smalheiser NR, Torvik VI. Complications in mammalian microRNA target prediction. Methods Mol Biol. 2006;342:115–27. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
