Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound
- PMID: 24476948
- PMCID: PMC4030780
- DOI: 10.1093/hmg/ddu038
Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound
Abstract
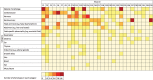
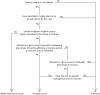
The genetic etiology of non-aneuploid fetal structural abnormalities is typically investigated by karyotyping and array-based detection of microscopically detectable rearrangements, and submicroscopic copy-number variants (CNVs), which collectively yield a pathogenic finding in up to 10% of cases. We propose that exome sequencing may substantially increase the identification of underlying etiologies. We performed exome sequencing on a cohort of 30 non-aneuploid fetuses and neonates (along with their parents) with diverse structural abnormalities first identified by prenatal ultrasound. We identified candidate pathogenic variants with a range of inheritance models, and evaluated these in the context of detailed phenotypic information. We identified 35 de novo single-nucleotide variants (SNVs), small indels, deletions or duplications, of which three (accounting for 10% of the cohort) are highly likely to be causative. These are de novo missense variants in FGFR3 and COL2A1, and a de novo 16.8 kb deletion that includes most of OFD1. In five further cases (17%) we identified de novo or inherited recessive or X-linked variants in plausible candidate genes, which require additional validation to determine pathogenicity. Our diagnostic yield of 10% is comparable to, and supplementary to, the diagnostic yield of existing microarray testing for large chromosomal rearrangements and targeted CNV detection. The de novo nature of these events could enable couples to be counseled as to their low recurrence risk. This study outlines the way for a substantial improvement in the diagnostic yield of prenatal genetic abnormalities through the application of next-generation sequencing.
© The Author 2014. Published by Oxford University Press.
Figures
References
-
- Springett A., Morris J.K. Congenital anomaly statistics. England and Wales. London: British Isles Network of Congenital Anomaly Registers; 2010.
-
- Hillman S.C., McMullan D.J., Hall G., Togneri F.S., James N., Maher E.J., Meller C.H., Williams D., Wapner R.J., Maher E.R., et al. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2013;41:610–620. - PubMed
-
- Valduga M., Philippe C., Bach Segura P., Thiebaugeorges O., Miton A., Beri M., Bonnet C., Nemos C., Foliguet B., Jonveaux P. A retrospective study by oligonucleotide array-CGH analysis in 50 fetuses with multiple malformations. Prenat. Diagn. 2010;30:333–341. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

