DNA-binding specificities of plant transcription factors and their potential to define target genes
- PMID: 24477691
- PMCID: PMC3926073
- DOI: 10.1073/pnas.1316278111
DNA-binding specificities of plant transcription factors and their potential to define target genes
Abstract
Transcription factors (TFs) regulate gene expression through binding to cis-regulatory specific sequences in the promoters of their target genes. In contrast to the genetic code, the transcriptional regulatory code is far from being deciphered and is determined by sequence specificity of TFs, combinatorial cooperation between TFs and chromatin competence. Here we addressed one of these determinants by characterizing the target sequence specificity of 63 plant TFs representing 25 families, using protein-binding microarrays. Remarkably, almost half of these TFs recognized secondary motifs, which in some cases were completely unrelated to the primary element. Analyses of coregulated genes and transcriptomic data from TFs mutants showed the functional significance of over 80% of all identified sequences and of at least one target sequence per TF. Moreover, combining the target sequence information with coexpression analysis we could predict the function of a TF as activator or repressor through a particular DNA sequence. Our data support the correlation between cis-regulatory elements and the sequence determined in vitro using the protein-binding microarray and provides a framework to explore regulatory networks in plants.
Keywords: Arabidopsis; cis-element; coregulation; regulatory network; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

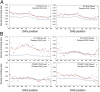

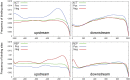
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

