Sterol regulatory element-binding protein (SREBP) cleavage regulates Golgi-to-endoplasmic reticulum recycling of SREBP cleavage-activating protein (SCAP)
- PMID: 24478315
- PMCID: PMC3953268
- DOI: 10.1074/jbc.M113.545699
Sterol regulatory element-binding protein (SREBP) cleavage regulates Golgi-to-endoplasmic reticulum recycling of SREBP cleavage-activating protein (SCAP)
Abstract
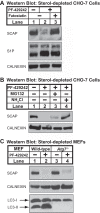
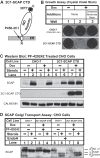
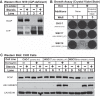
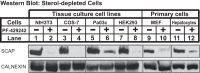
Sterol regulatory element-binding protein (SREBP) transcription factors are central regulators of cellular lipogenesis. Release of membrane-bound SREBP requires SREBP cleavage-activating protein (SCAP) to escort SREBP from the endoplasmic reticulum (ER) to the Golgi for cleavage by site-1 and site-2 proteases. SCAP then recycles to the ER for additional rounds of SREBP binding and transport. Mechanisms regulating ER-to-Golgi transport of SCAP-SREBP are understood in molecular detail, but little is known about SCAP recycling. Here, we have demonstrated that SCAP Golgi-to-ER transport requires cleavage of SREBP at site-1. Reductions in SREBP cleavage lead to SCAP degradation in lysosomes, providing additional negative feedback control to the SREBP pathway. Current models suggest that SREBP plays a passive role prior to cleavage. However, we show that SREBP actively prevents premature recycling of SCAP-SREBP until initiation of SREBP cleavage. SREBP regulates SCAP in human cells and yeast, indicating that this is an ancient regulatory mechanism.
Keywords: Cholesterol; Lipids; Lysosomes; Protein Degradation; Recycling; SCAP; SREBP; Transcription Factors.
Figures

References
-
- Hachinski V., Graffagnino C., Beaudry M., Bernier G., Buck C., Donner A., Spence J. D., Doig G., Wolfe B. M. (1996) Lipids and stroke: a paradox resolved. Arch. Neurol. 53, 303–308 - PubMed
-
- Rawson R. B., DeBose-Boyd R., Goldstein J. L., Brown M. S. (1999) Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274, 28549–28556 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

