The cell biology of the endocytic system from an evolutionary perspective
- PMID: 24478384
- PMCID: PMC3970418
- DOI: 10.1101/cshperspect.a016998
The cell biology of the endocytic system from an evolutionary perspective
Abstract
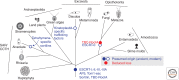
Evolutionary cell biology can afford an interdisciplinary comparative view that gives insights into both the functioning of modern cells and the origins of cellular systems, including the endocytic organelles. Here, we explore several recent evolutionary cell biology studies, highlighting investigations into the origin and diversity of endocytic systems in eukaryotes. Beginning with a brief overview of the eukaryote tree of life, we show how understanding the endocytic machinery in a select, but diverse, array of organisms provides insights into endocytic system origins and predicts the likely configuration in the last eukaryotic common ancestor (LECA). Next, we consider three examples in which a comparative approach yielded insight into the function of modern cellular systems. First, using ESCRT-0 as an example, we show how comparative cell biology can discover both lineage-specific novelties (ESCRT-0) as well as previously ignored ancient proteins (Tom1), likely of both evolutionary and functional importance. Second, we highlight the power of comparative cell biology for discovery of previously ignored but potentially ancient complexes (AP5). Finally, using examples from ciliates and trypanosomes, we show that not all organisms possess canonical endocytic pathways, but instead likely evolved lineage-specific mechanisms. Drawing from these case studies, we conclude that a comparative approach is a powerful strategy for advancing knowledge about the general mechanisms and functions of endocytic systems.
Figures


References
-
- Adung’a VO, Gadelha C, Field MC 2013. Proteomic analysis of clathrin interactions in trypanosomes reveals dynamic evolution of endocytosis. Traffic 14: 440–457 - PubMed
-
- Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F 2009. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic 10: 161–171 - PubMed
-
- Bogan JS 2012. Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem 81: 507–532 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
