Advances in targeting the vacuolar proton-translocating ATPase (V-ATPase) for anti-fungal therapy
- PMID: 24478704
- PMCID: PMC3902353
- DOI: 10.3389/fphar.2014.00004
Advances in targeting the vacuolar proton-translocating ATPase (V-ATPase) for anti-fungal therapy
Abstract
Vacuolar proton-translocating ATPase (V-ATPase) is a membrane-bound, multi-subunit enzyme that uses the energy of ATP hydrolysis to pump protons across membranes. V-ATPase activity is critical for pH homeostasis and organelle acidification as well as for generation of the membrane potential that drives secondary transporters and cellular metabolism. V-ATPase is highly conserved across species and is best characterized in the model fungus Saccharomyces cerevisiae. However, recent studies in mammals have identified significant alterations from fungi, particularly in the isoform composition of the 14 subunits and in the regulation of complex disassembly. These differences could be exploited for selectivity between fungi and humans and highlight the potential for V-ATPase as an anti-fungal drug target. Candida albicans is a major human fungal pathogen and causes fatality in 35% of systemic infections, even with anti-fungal treatment. The pathogenicity of C. albicans correlates with environmental, vacuolar, and cytoplasmic pH regulation, and V-ATPase appears to play a fundamental role in each of these processes. Genetic loss of V-ATPase in pathogenic fungi leads to defective virulence, and a comprehensive picture of the mechanisms involved is emerging. Recent studies have explored the practical utility of V-ATPase as an anti-fungal drug target in C. albicans, including pharmacological inhibition, azole therapy, and targeting of downstream pathways. This overview will discuss these studies as well as hypothetical ways to target V-ATPase and novel high-throughput methods for use in future drug discovery screens.
Keywords: C. albicans virulence; anti-fungal target; fungal V-ATPase; pH homeostasis; vacuolar acidification; vacuolar proton pump.
Figures
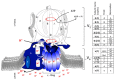
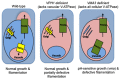
Similar articles
-
Vacuolar proton-translocating ATPase is required for antifungal resistance and virulence of Candida glabrata.PLoS One. 2019 Jan 23;14(1):e0210883. doi: 10.1371/journal.pone.0210883. eCollection 2019. PLoS One. 2019. PMID: 30673768 Free PMC article.
-
Deletion of vacuolar proton-translocating ATPase V(o)a isoforms clarifies the role of vacuolar pH as a determinant of virulence-associated traits in Candida albicans.J Biol Chem. 2013 Mar 1;288(9):6190-201. doi: 10.1074/jbc.M112.426197. Epub 2013 Jan 11. J Biol Chem. 2013. PMID: 23316054 Free PMC article.
-
Candida albicans VMA3 is necessary for V-ATPase assembly and function and contributes to secretion and filamentation.Eukaryot Cell. 2013 Oct;12(10):1369-82. doi: 10.1128/EC.00118-13. Epub 2013 Aug 2. Eukaryot Cell. 2013. PMID: 23913543 Free PMC article.
-
Reciprocal Regulation of V-ATPase and Glycolytic Pathway Elements in Health and Disease.Front Physiol. 2019 Feb 15;10:127. doi: 10.3389/fphys.2019.00127. eCollection 2019. Front Physiol. 2019. PMID: 30828305 Free PMC article. Review.
-
The V-type H+ ATPase: molecular structure and function, physiological roles and regulation.J Exp Biol. 2006 Feb;209(Pt 4):577-89. doi: 10.1242/jeb.02014. J Exp Biol. 2006. PMID: 16449553 Review.
Cited by
-
FcRav2, a gene with a ROGDI domain involved in Fusarium head blight and crown rot on durum wheat caused by Fusarium culmorum.Mol Plant Pathol. 2018 Mar;19(3):677-688. doi: 10.1111/mpp.12551. Epub 2017 May 31. Mol Plant Pathol. 2018. PMID: 28322011 Free PMC article.
-
Vacuolar proton-translocating ATPase is required for antifungal resistance and virulence of Candida glabrata.PLoS One. 2019 Jan 23;14(1):e0210883. doi: 10.1371/journal.pone.0210883. eCollection 2019. PLoS One. 2019. PMID: 30673768 Free PMC article.
-
The Role of Secretory Pathways in Candida albicans Pathogenesis.J Fungi (Basel). 2020 Feb 24;6(1):26. doi: 10.3390/jof6010026. J Fungi (Basel). 2020. PMID: 32102426 Free PMC article. Review.
-
Attenuation of Candida albicans virulence with focus on disruption of its vacuole functions.J Oral Microbiol. 2014 Apr 1;6. doi: 10.3402/jom.v6.23898. eCollection 2014. J Oral Microbiol. 2014. PMID: 24765242 Free PMC article. Review.
-
Rotation Mechanism of Molecular Motor V1-ATPase Studied by Multiscale Molecular Dynamics Simulation.Biophys J. 2017 Mar 14;112(5):911-920. doi: 10.1016/j.bpj.2017.01.029. Biophys J. 2017. PMID: 28297650 Free PMC article.
References
-
- Bowman E. J., O’Neill F. J., Bowman B. J. (1997). Mutations of pma-1, the gene encoding the plasma membrane H+-ATPase of Neurospora crassa, suppress inhibition of growth by concanamycin A, a specific inhibitor of vacuolar ATPases. J. Biol. Chem. 272 14776–14786 10.1074/jbc.272.23.14776 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

