Analysis of the Lotus japonicus nuclear pore NUP107-160 subcomplex reveals pronounced structural plasticity and functional redundancy
- PMID: 24478780
- PMCID: PMC3897872
- DOI: 10.3389/fpls.2013.00552
Analysis of the Lotus japonicus nuclear pore NUP107-160 subcomplex reveals pronounced structural plasticity and functional redundancy
Abstract
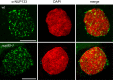
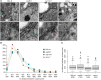
Mutations in the Lotus japonicus nucleoporin genes, NUP85, NUP133, and NENA (SEH1), lead to defects in plant-microbe symbiotic signaling. The homologous proteins in yeast and vertebrates are part of the conserved NUP84/NUP107-160 subcomplex, which is an essential component of the nuclear pore scaffold and has a pivotal role in nuclear pore complex (NPC) assembly. Loss and down-regulation of NUP84/NUP107-160 members has previously been correlated with a variety of growth and molecular defects, however, in L. japonicus only surprisingly specific phenotypes have been reported. We investigated whether Lotus nup85, nup133, and nena mutants exhibit general defects in NPC composition and distribution. Whole mount immunolocalization confirmed a typical nucleoporin-like localization for NUP133, which was unchanged in the nup85-1 mutant. Severe NPC clustering and aberrations in the nuclear envelope have been reported for Saccharomyces cerevisiae nup85 and nup133 mutants. However, upon transmission electron microscopy analysis of L. japonicus nup85, nup133 and nena, we detected only a slight reduction in the average distances between neighboring NPCs in nup133. Using quantitative immunodetection on protein-blots we observed that loss of individual nucleoporins affected the protein levels of other NUP107-160 complex members. Unlike the single mutants, nup85/nup133 double mutants exhibited severe temperature dependent growth and developmental defects, suggesting that the loss of more than one NUP107-160 member affects basal functions of the NPC.
Keywords: Lotus japonicus; NUP107-160 subcomplex; nuclear pore complex; nucleoporins; plant nucleus.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

