Variant calling in low-coverage whole genome sequencing of a Native American population sample
- PMID: 24479562
- PMCID: PMC3914019
- DOI: 10.1186/1471-2164-15-85
Variant calling in low-coverage whole genome sequencing of a Native American population sample
Abstract
Background: The reduction in the cost of sequencing a human genome has led to the use of genotype sampling strategies in order to impute and infer the presence of sequence variants that can then be tested for associations with traits of interest. Low-coverage Whole Genome Sequencing (WGS) is a sampling strategy that overcomes some of the deficiencies seen in fixed content SNP array studies. Linkage-disequilibrium (LD) aware variant callers, such as the program Thunder, may provide a calling rate and accuracy that makes a low-coverage sequencing strategy viable.
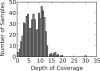
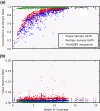
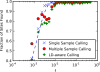
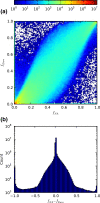
Results: We examined the performance of an LD-aware variant calling strategy in a population of 708 low-coverage whole genome sequences from a community sample of Native Americans. We assessed variant calling through a comparison of the sequencing results to genotypes measured in 641 of the same subjects using a fixed content first generation exome array. The comparison was made using the variant calling routines GATK Unified Genotyper program and the LD-aware variant caller Thunder. Thunder was found to improve concordance in a coverage dependent fashion, while correctly calling nearly all of the common variants as well as a high percentage of the rare variants present in the sample.
Conclusions: Low-coverage WGS is a strategy that appears to collect genetic information intermediate in scope between fixed content genotyping arrays and deep-coverage WGS. Our data suggests that low-coverage WGS is a viable strategy with a greater chance of discovering novel variants and associations than fixed content arrays for large sample association analyses.
Figures

References
-
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;15:64–69. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

