Fitting CRISPR-associated Cas3 into the helicase family tree
- PMID: 24480304
- PMCID: PMC3984625
- DOI: 10.1016/j.sbi.2014.01.001
Fitting CRISPR-associated Cas3 into the helicase family tree
Abstract
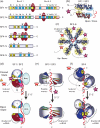
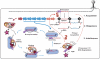
Helicases utilize NTPs to modulate their binding to nucleic acids and many of these enzymes also unwind DNA or RNA duplexes in an NTP-dependent fashion. These proteins are phylogenetically related but functionally diverse, with essential roles in virtually all aspects of nucleic acid metabolism. A new class of helicases associated with RNA-guided adaptive immune systems in bacteria and archaea has recently been identified. Prokaryotes acquire resistance to invading genetic parasites by integrating short fragments of foreign nucleic acids into repetitive loci in the host chromosome known as CRISPRs (Clustered Regularly Interspaced Short Palindromic Repeats). CRISPR-associated gene 3 (cas3) encodes a conserved helicase protein that is essential for phage defense. Here we review recent advances in Cas3 biology, and provide a new phylogenetic framework that positions Cas3 in the helicase family tree. We anticipate that this Cas3 phylogeny will guide future biochemical and structural studies.
Copyright © 2014. Published by Elsevier Ltd.
Figures

Similar articles
-
Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16359-64. doi: 10.1073/pnas.1410806111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368186 Free PMC article.
-
Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation.Mol Cell. 2016 Sep 1;63(5):852-64. doi: 10.1016/j.molcel.2016.07.011. Epub 2016 Aug 18. Mol Cell. 2016. PMID: 27546790
-
CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6618-23. doi: 10.1073/pnas.1405079111. Epub 2014 Apr 18. Proc Natl Acad Sci U S A. 2014. PMID: 24748111 Free PMC article.
-
Cas3 Protein-A Review of a Multi-Tasking Machine.Genes (Basel). 2020 Feb 18;11(2):208. doi: 10.3390/genes11020208. Genes (Basel). 2020. PMID: 32085454 Free PMC article. Review.
-
Unravelling the structural and mechanistic basis of CRISPR-Cas systems.Nat Rev Microbiol. 2014 Jul;12(7):479-92. doi: 10.1038/nrmicro3279. Epub 2014 Jun 9. Nat Rev Microbiol. 2014. PMID: 24909109 Free PMC article. Review.
Cited by
-
Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system.Nat Commun. 2016 Oct 3;7:12853. doi: 10.1038/ncomms12853. Nat Commun. 2016. PMID: 27694798 Free PMC article.
-
Metal Dependence and Functional Diversity of Type I Cas3 Nucleases.Biochemistry. 2022 Mar 1;61(5):327-338. doi: 10.1021/acs.biochem.1c00779. Epub 2022 Feb 21. Biochemistry. 2022. PMID: 35184547 Free PMC article.
-
Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.Nat Struct Mol Biol. 2014 Sep;21(9):771-7. doi: 10.1038/nsmb.2875. Epub 2014 Aug 17. Nat Struct Mol Biol. 2014. PMID: 25132177 Free PMC article.
-
DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus.Nucleic Acids Res. 2015 Dec 2;43(21):10353-63. doi: 10.1093/nar/gkv1140. Epub 2015 Oct 30. Nucleic Acids Res. 2015. PMID: 26519471 Free PMC article.
-
Current and future prospects for CRISPR-based tools in bacteria.Biotechnol Bioeng. 2016 May;113(5):930-43. doi: 10.1002/bit.25851. Epub 2015 Oct 27. Biotechnol Bioeng. 2016. PMID: 26460902 Free PMC article. Review.
References
-
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. [This review provides a comprehensive summary of the structure, function and nomenclature of helicases.] - PubMed
-
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annual Review of Biophysics. 2008;37:317–336. - PubMed
-
- Steimer L, Klostermeier D. RNA helicases in infection and disease. RNA Biology. 2012;9:751–771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

