Structural studies of the spliceosome: zooming into the heart of the machine
- PMID: 24480332
- PMCID: PMC4045393
- DOI: 10.1016/j.sbi.2013.12.002
Structural studies of the spliceosome: zooming into the heart of the machine
Abstract
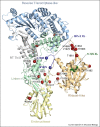
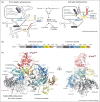
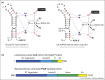
Spliceosomes are large, dynamic ribonucleoprotein complexes that catalyse the removal of introns from messenger RNA precursors via a two-step splicing reaction. The recent crystal structure of Prp8 has revealed Reverse Transcriptase-like, Linker and Endonuclease-like domains. The intron branch-point cross-link with the Linker domain of Prp8 in active spliceosomes and together with suppressors of 5' and 3' splice site mutations this unambiguously locates the active site cavity. Structural and mechanistic similarities with group II self-splicing introns have encouraged the notion that the spliceosome is at heart a ribozyme, and recently the ligands for two catalytic magnesium ions were identified within U6 snRNA. They position catalytic divalent metal ions in the same way as Domain V of group II intron RNA, suggesting that the spliceosome and group II intron use the same catalytic mechanisms.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Cordin O., Hahn D., Beggs J.D. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol. 2012;24:431–438. - PubMed
-
- Newman A.J., Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. - PubMed
-
- Lührmann R., Stark H. Structural mapping of spliceosomes by electron microscopy. Curr Opin Struct Biol. 2009;19:96–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

