Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency
- PMID: 24480542
- PMCID: PMC3958305
- DOI: 10.1002/emmm.201303573
Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency
Abstract
The molecular basis of a significant number of cases of isolated growth hormone deficiency remains unknown. We describe three sisters affected with severe isolated growth hormone deficiency and pituitary hypoplasia caused by biallelic mutations in the RNPC3 gene, which codes for a minor spliceosome protein required for U11/U12 small nuclear ribonucleoprotein (snRNP) formation and splicing of U12-type introns. We found anomalies in U11/U12 di-snRNP formation and in splicing of multiple U12-type introns in patient cells. Defective transcripts include preprohormone convertases SPCS2 and SPCS3 and actin-related ARPC5L genes, which are candidates for the somatotroph-restricted dysfunction. The reported novel mechanism for familial growth hormone deficiency demonstrates that general mRNA processing defects of the minor spliceosome can lead to very narrow tissue-specific consequences.
Figures
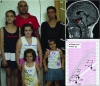
Photograph of family members showing the marked short stature of the three affected girls compared to unaffected relatives, along with other typical features of GHD such as frontal bossing and cherubic face. Ages are shown over each girl, with the height scale on the right.
Midline sagittal section of the brain MRI of one proband (IGHD-02). A hypoplastic pituitary is evident (red arrow) with no other brain anomaly.
Growth chart including the anthropometric data of the three affected siblings, bone age and height showing delayed and slow basal growth and an excellent response to GH replacement therapy (rGH).
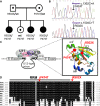
Family pedigree showing that all three affected girls are compound heterozygotes for the RNPC3 mutations shown under each symbol while the unaffected parents and sister are heterozygous carriers for one mutation.
Sequencing of the RT-PCR product of RNPC3 from blood RNA of the eldest proband showing the p.P474T (top) and p.R502X mutations in the same amplicon, with relatively decreased expression of the non-sense carrying allele (p.R502X).
Schematic representation of the function of the RNPC3 gene product, U11/U12-65K protein, in RNA splicing. The 65K protein is part of a molecular bridge that links U11 and U12 snRNPs in the intron recognition complex. The mutations may disturb binding of U12 snRNA to 65K protein. Inset indicates the positions of the mutations (red circles) in the 3D structure of the second RRM of the 65K protein determined by X-ray crystallography (Protein Data Bank Identification, PDB-ID: 3EGN).
Sequence alignment of the 65K protein second RRM of multiple species using ClustalW algorithm. The mutated residues (red arrows) are highly conserved phylogenetically. (HSA: Homo sapiens; PTR: Pan troglodytes; MMU: Macaca mulatta; CAN: Canis lupus; BTA: Bos taurus; MM: Mus musculus; RN: Rattus norvegicus; GG: Gallus gallus; DR: Danio rerio; AT: Arabidopsis thaliana; OS: Oryza sativa).
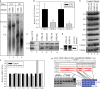
Native gel analysis of U11 and U12 snRNP complexes in nuclear extracts.
U12 snRNP pull-down assay measuring the level of U11 snRNP associated with U12 snRNP. Three technical replicates were used for each pulldown. Student's t test, **P = 0.02, ***P = 0.01, n = 3.
Representative gel of data in panel B.
Western blot analysis of control and patient nuclear extracts using antibodies targeted to U11/U12-65K protein. Purified U11/U12 di-snRNP was used as control in lane 3.
Northern blot analysis of expression levels of spliceosomal snRNAs in lymphoblastoid cell lines from two patients (IGHD-01 and IGHD-02) and two controls (CTRL-01 = LCL422 and CTRL-02 = LCL452). Total RNA was extracted from each cell line and individual snRNAs detected by Northern blotting.
Quantification of the snRNAs in D. Data represents mean values of two patient and two control datasets.
Transcription profiles of the SCPS2 gene. RNAseq and RT-PCR obtained concordant results showing relatively poor U12-type splicing in patients with increased intron retention (U12 and flanking U2 introns), along with alternative (aberrant) U2-type splicing (˜30% of transcripts). The alternative transcripts are barely present in controls or heterozygous carriers. The band in genomic DNA lane (gDNA) is derived from a processed pseudogene on chromosome 1. Verification of the appropriate content (real processed mRNA in the cDNA products and processed pseudogene in the genomic amplification) was performed by sequencing.
References
-
- Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for Geneticists. Chichester, UK: John Wiley ' Sons Ltd; 2003.
-
- Boulisfane N, Choleza M, Rage F, Soret J, Bordonné R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet. 2011;20:641–648. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

