PIFs: systems integrators in plant development
- PMID: 24481072
- PMCID: PMC3963594
- DOI: 10.1105/tpc.113.120857
PIFs: systems integrators in plant development
Abstract
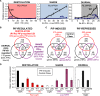
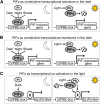
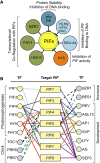
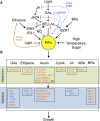
Phytochrome-interacting factors (PIFs) are members of the Arabidopsis thaliana basic helix-loop-helix family of transcriptional regulators that interact specifically with the active Pfr conformer of phytochrome (phy) photoreceptors. PIFs are central regulators of photomorphogenic development that act to promote stem growth, and this activity is reversed upon interaction with phy in response to light. Recently, significant progress has been made in defining the transcriptional networks directly regulated by PIFs, as well as the convergence of other signaling pathways on the PIFs to modulate growth. Here, we summarize and highlight these findings in the context of PIFs acting as integrators of light and other signals. We discuss progress in our understanding of the transcriptional and posttranslational regulation of PIFs that illustrates the integration of light with hormonal pathways and the circadian clock, and we review seedling hypocotyl growth as a paradigm of PIFs acting at the interface of these signals. Based on these advances, PIFs are emerging as required factors for growth, acting as central components of a regulatory node that integrates multiple internal and external signals to optimize plant development.
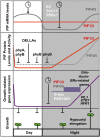
Figures

References
-
- Alabadí D., Gallego-Bartolomé J., Orlando L., García-Cárcel L., Rubio V., Martínez C., Frigerio M., Iglesias-Pedraz J.M., Espinosa A., Deng X.W., Blázquez M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53: 324–335 - PubMed
-
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

