The UV-B photoreceptor UVR8: from structure to physiology
- PMID: 24481075
- PMCID: PMC3963570
- DOI: 10.1105/tpc.113.119446
The UV-B photoreceptor UVR8: from structure to physiology
Abstract
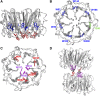
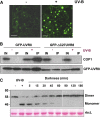
Low doses of UV-B light (280 to 315 nm) elicit photomorphogenic responses in plants that modify biochemical composition, photosynthetic competence, morphogenesis, and defense. UV RESISTANCE LOCUS8 (UVR8) mediates photomorphogenic responses to UV-B by regulating transcription of a set of target genes. UVR8 differs from other known photoreceptors in that it uses specific Trp amino acids instead of a prosthetic chromophore for light absorption during UV-B photoreception. Absorption of UV-B dissociates the UVR8 dimer into monomers, initiating signal transduction through interaction with CONSTITUTIVELY PHOTOMORPHOGENIC1. However, much remains to be learned about the physiological role of UVR8 and its interaction with other signaling pathways, the molecular mechanism of UVR8 photoreception, how the UVR8 protein initiates signaling, how it is regulated, and how UVR8 regulates transcription of its target genes.
Figures


References
-
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 - PubMed
-
- Ballaré C.L., Barnes P.W., Flint S.D. (1995). Inhibition of hypocotyl elongation by ultraviolet-B radiation in de-etiolating tomato seedlings. 1. The photoreceptor. Physiol. Plant. 93: 584–592
-
- Belles-Boix E., Babiychuk E., Van Montagu M., Inzé D., Kushnir S. (2000). CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 482: 19–24 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

