Why are male malaria parasites in such a rush?: Sex-specific evolution and host-parasite interactions
- PMID: 24481180
- PMCID: PMC4183958
- DOI: 10.1093/emph/eos003
Why are male malaria parasites in such a rush?: Sex-specific evolution and host-parasite interactions
Abstract
Background: Disease-causing organisms are notorious for fast rates of molecular evolution and the ability to adapt rapidly to changes in their ecology. Sex plays a key role in evolution, and recent studies, in humans and other multicellular organisms, document that genes expressed principally or exclusively in males exhibit the fastest rates of adaptive evolution. However, despite the importance of sexual reproduction for many unicellular taxa, sex-biased gene expression and its evolutionary implications have been overlooked.
Methods: We analyse genomic data from multiple malaria parasite (Plasmodium) species and proteomic data sets from different parasite life cycle stages.
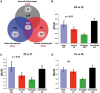
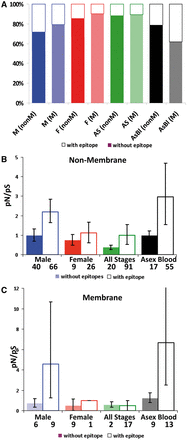
Results: The accelerated evolution of male-biased genes has only been examined in multicellular taxa, but our analyses reveal that accelerated evolution in genes with male-specific expression is also a feature of unicellular organisms. This 'fast-male' evolution is adaptive and likely facilitated by the male-biased sex ratio of gametes in the mating pool. Furthermore, we propose that the exceptional rates of evolution we observe are driven by interactions between males and host immune responses.
Conclusions: We reveal a novel form of host-parasite coevolution that enables parasites to evade host immune responses that negatively impact upon fertility. The identification of parasite genes with accelerated evolution has important implications for the identification of drug and vaccine targets. Specifically, vaccines targeting males will be more vulnerable to parasite evolution than those targeting females or both sexes.
Keywords: Plasmodium; gene expression; host–parasite coevolution; sex-specific selection.
Figures

Similar articles
-
Sex and death: the effects of innate immune factors on the sexual reproduction of malaria parasites.PLoS Pathog. 2011 Mar;7(3):e1001309. doi: 10.1371/journal.ppat.1001309. Epub 2011 Mar 3. PLoS Pathog. 2011. PMID: 21408620 Free PMC article.
-
The ecology of fish parasites with particular reference to helminth parasites and their salmonid fish hosts in Welsh rivers: a review of some of the central questions.Adv Parasitol. 2002;52:1-154. doi: 10.1016/s0065-308x(02)52011-x. Adv Parasitol. 2002. PMID: 12521260 Review.
-
Lineage-specific positive selection at the merozoite surface protein 1 (msp1) locus of Plasmodium vivax and related simian malaria parasites.BMC Evol Biol. 2010 Feb 19;10:52. doi: 10.1186/1471-2148-10-52. BMC Evol Biol. 2010. PMID: 20167126 Free PMC article.
-
Host-Malaria Parasite Interactions and Impacts on Mutual Evolution.Front Cell Infect Microbiol. 2020 Oct 27;10:587933. doi: 10.3389/fcimb.2020.587933. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194831 Free PMC article. Review.
-
Adaptive plasticity in the gametocyte conversion rate of malaria parasites.PLoS Pathog. 2018 Nov 14;14(11):e1007371. doi: 10.1371/journal.ppat.1007371. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30427935 Free PMC article.
Cited by
-
Checks and balances? DNA replication and the cell cycle in Plasmodium.Parasit Vectors. 2018 Mar 27;11(1):216. doi: 10.1186/s13071-018-2800-1. Parasit Vectors. 2018. PMID: 29587837 Free PMC article. Review.
-
The private life of malaria parasites: Strategies for sexual reproduction.Mol Biochem Parasitol. 2021 Jul;244:111375. doi: 10.1016/j.molbiopara.2021.111375. Epub 2021 May 20. Mol Biochem Parasitol. 2021. PMID: 34023299 Free PMC article. Review.
-
Discordant population histories of host and its parasite: A role for ecological permeability of extreme environment?PLoS One. 2017 Apr 10;12(4):e0175286. doi: 10.1371/journal.pone.0175286. eCollection 2017. PLoS One. 2017. PMID: 28394904 Free PMC article.
-
War and peace: social interactions in infections.Philos Trans R Soc Lond B Biol Sci. 2014 Mar 31;369(1642):20130365. doi: 10.1098/rstb.2013.0365. Print 2014 May 19. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 24686936 Free PMC article. Review.
-
Biology of Plasmodium falciparum gametocyte sex ratio and implications in malaria parasite transmission.Malar J. 2019 Mar 12;18(1):70. doi: 10.1186/s12936-019-2707-0. Malar J. 2019. PMID: 30866941 Free PMC article. Review.
References
-
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–98. - PubMed
-
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, et al. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–5. - PubMed
-
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–9. - PubMed
-
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–9. - PubMed
-
- Good JM, Nachman MW. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol Biol Evol. 2005;22:1044–52. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

