BMAL1-dependent regulation of the mTOR signaling pathway delays aging
- PMID: 24481314
- PMCID: PMC3927809
- DOI: 10.18632/aging.100633
BMAL1-dependent regulation of the mTOR signaling pathway delays aging
Abstract
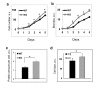
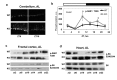
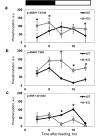
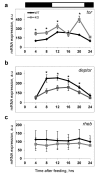
The circadian clock, an internal time-keeping system, has been linked with control of aging, but molecular mechanisms of regulation are not known. BMAL1 is a transcriptional factor and core component of the circadian clock; BMAL1 deficiency is associated with premature aging and reduced lifespan. Here we report that activity of mammalian Target of Rapamycin Complex 1 (mTORC1) is increased upon BMAL1 deficiency both in vivo and in cell culture. Increased mTOR signaling is associated with accelerated aging; in accordance with that, treatment with the mTORC1 inhibitor rapamycin increased lifespan of Bmal1-/- mice by 50%. Our data suggest that BMAL1 is a negative regulator of mTORC1 signaling. We propose that the circadian clock controls the activity of the mTOR pathway through BMAL1-dependent mechanisms and this regulation is important for control of aging and metabolism.
Conflict of interest statement
The authors of this paper declare no conflict of interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

