uPEPperoni: an online tool for upstream open reading frame location and analysis of transcript conservation
- PMID: 24484385
- PMCID: PMC3914846
- DOI: 10.1186/1471-2105-15-36
uPEPperoni: an online tool for upstream open reading frame location and analysis of transcript conservation
Abstract
Background: Several small open reading frames located within the 5' untranslated regions of mRNAs have recently been shown to be translated. In humans, about 50% of mRNAs contain at least one upstream open reading frame representing a large resource of coding potential. We propose that some upstream open reading frames encode peptides that are functional and contribute to proteome complexity in humans and other organisms. We use the term uPEPs to describe peptides encoded by upstream open reading frames.
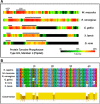
Results: We have developed an online tool, termed uPEPperoni, to facilitate the identification of putative bioactive peptides. uPEPperoni detects conserved upstream open reading frames in eukaryotic transcripts by comparing query nucleotide sequences against mRNA sequences within the NCBI RefSeq database. The algorithm first locates the main coding sequence and then searches for open reading frames 5' to the main start codon which are subsequently analysed for conservation. uPEPperoni also determines the substitution frequency for both the upstream open reading frames and the main coding sequence. In addition, the uPEPperoni tool produces sequence identity heatmaps which allow rapid visual inspection of conserved regions in paired mRNAs.
Conclusions: uPEPperoni features user-nominated settings including, nucleotide match/mismatch, gap penalties, Ka/Ks ratios and output mode. The heatmap output shows levels of identity between any two sequences and provides easy recognition of conserved regions. Furthermore, this web tool allows comparison of evolutionary pressures acting on the upstream open reading frame against other regions of the mRNA. Additionally, the heatmap web applet can also be used to visualise the degree of conservation in any pair of sequences. uPEPperoni is freely available on an interactive web server at http://upep-scmb.biosci.uq.edu.au.
Figures


Similar articles
-
Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides.BMC Genomics. 2006 Jan 26;7:16. doi: 10.1186/1471-2164-7-16. BMC Genomics. 2006. PMID: 16438715 Free PMC article.
-
[Potential open reading frames within 5'-untranslated regions of eukaryotic mRNAs].Biofizika. 2006 Jul-Aug;51(4):615-21. Biofizika. 2006. PMID: 16909838 Russian.
-
Diversity of translation start sites may define increased complexity of the human short ORFeome.Mol Cell Proteomics. 2007 Jun;6(6):1000-6. doi: 10.1074/mcp.M600297-MCP200. Epub 2007 Feb 21. Mol Cell Proteomics. 2007. PMID: 17317662
-
Translational control by 5'-untranslated regions of eukaryotic mRNAs.Science. 2016 Jun 17;352(6292):1413-6. doi: 10.1126/science.aad9868. Science. 2016. PMID: 27313038 Free PMC article. Review.
-
Polycistronic peptide coding genes in eukaryotes--how widespread are they?Brief Funct Genomic Proteomic. 2009 Jan;8(1):68-74. doi: 10.1093/bfgp/eln054. Epub 2008 Dec 12. Brief Funct Genomic Proteomic. 2009. PMID: 19074495 Review.
Cited by
-
Death of a dogma: eukaryotic mRNAs can code for more than one protein.Nucleic Acids Res. 2016 Jan 8;44(1):14-23. doi: 10.1093/nar/gkv1218. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578573 Free PMC article. Review.
-
Recent advances in improving yield and immunity through transcription factor engineering.J Integr Plant Biol. 2025 Aug;67(8):2005-2027. doi: 10.1111/jipb.13932. Epub 2025 May 21. J Integr Plant Biol. 2025. PMID: 40396540 Free PMC article. Review.
-
A survey of experimental and computational identification of small proteins.Brief Bioinform. 2024 May 23;25(4):bbae345. doi: 10.1093/bib/bbae345. Brief Bioinform. 2024. PMID: 39007598 Free PMC article. Review.
-
Functionally Significant Features in the 5' Untranslated Region of the ABCA1 Gene and Their Comparison in Vertebrates.Cells. 2019 Jun 21;8(6):623. doi: 10.3390/cells8060623. Cells. 2019. PMID: 31234415 Free PMC article.
-
Pervasive translation of small open reading frames in plant long non-coding RNAs.Front Plant Sci. 2022 Oct 24;13:975938. doi: 10.3389/fpls.2022.975938. eCollection 2022. Front Plant Sci. 2022. PMID: 36352887 Free PMC article. Review.
References
-
- Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

