The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation
- PMID: 24484546
- PMCID: PMC3928079
- DOI: 10.1186/1471-2164-15-92
The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation
Abstract
Background: Chromatin architectural proteins interact with nucleosomes to modulate chromatin accessibility and higher-order chromatin structure. While these proteins are almost certainly important for gene regulation they have been studied far less than the core histone proteins.
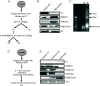
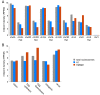
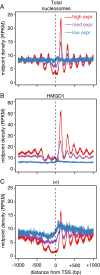
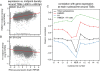
Results: Here we describe the genomic distributions and functional roles of two chromatin architectural proteins: histone H1 and the high mobility group protein HMGD1 in Drosophila S2 cells. Using ChIP-seq, biochemical and gene specific approaches, we find that HMGD1 binds to highly accessible regulatory chromatin and active promoters. In contrast, H1 is primarily associated with heterochromatic regions marked with repressive histone marks. We find that the ratio of HMGD1 to H1 binding is a better predictor of gene activity than either protein by itself, which suggests that reciprocal binding between these proteins is important for gene regulation. Using knockdown experiments, we show that HMGD1 and H1 affect the occupancy of the other protein, change nucleosome repeat length and modulate gene expression.
Conclusion: Collectively, our data suggest that dynamic and mutually exclusive binding of H1 and HMGD1 to nucleosomes and their linker sequences may control the fluid chromatin structure that is required for transcriptional regulation. This study provides a framework to further study the interplay between chromatin architectural proteins and epigenetics in gene regulation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

