voom: Precision weights unlock linear model analysis tools for RNA-seq read counts
- PMID: 24485249
- PMCID: PMC4053721
- DOI: 10.1186/gb-2014-15-2-r29
voom: Precision weights unlock linear model analysis tools for RNA-seq read counts
Abstract
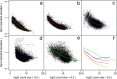
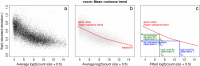
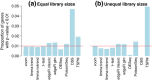
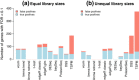
New normal linear modeling strategies are presented for analyzing read counts from RNA-seq experiments. The voom method estimates the mean-variance relationship of the log-counts, generates a precision weight for each observation and enters these into the limma empirical Bayes analysis pipeline. This opens access for RNA-seq analysts to a large body of methodology developed for microarrays. Simulation studies show that voom performs as well or better than count-based RNA-seq methods even when the data are generated according to the assumptions of the earlier methods. Two case studies illustrate the use of linear modeling and gene set testing methods.
Figures
References
-
- Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

