Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis
- PMID: 24485453
- PMCID: PMC3951122
- DOI: 10.1016/j.cell.2013.12.039
Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis
Abstract
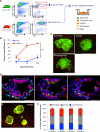
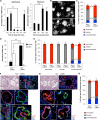
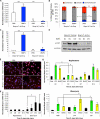
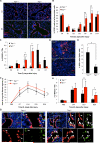
Lung stem cells are instructed to produce lineage-specific progeny through unknown factors in their microenvironment. We used clonal 3D cocultures of endothelial cells and distal lung stem cells, bronchioalveolar stem cells (BASCs), to probe the instructive mechanisms. Single BASCs had bronchiolar and alveolar differentiation potential in lung endothelial cell cocultures. Gain- and loss-of-function experiments showed that BMP4-Bmpr1a signaling triggers calcineurin/NFATc1-dependent expression of thrombospondin-1 (Tsp1) in lung endothelial cells to drive alveolar lineage-specific BASC differentiation. Tsp1 null mice exhibited defective alveolar injury repair, confirming a crucial role for the BMP4-NFATc1-TSP1 axis in lung epithelial differentiation and regeneration in vivo. Discovery of this pathway points to methods to direct the derivation of specific lung epithelial lineages from multipotent cells. These findings elucidate a pathway that may be a critical target in lung diseases and provide tools to understand the mechanisms of respiratory diseases at the single-cell level.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Aso Y, Yoneda K, Kikkawa Y. Morphologic and biochemical study of pulmonary changes induced by bleomycin in mice. Lab Invest. 1976;35:558–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

