UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma
- PMID: 24486181
- PMCID: PMC3951208
- DOI: 10.1016/j.ccr.2014.01.003
UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma
Abstract
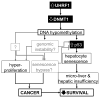
Ubiquitin-like with PHD and RING finger domains 1 (UHRF1) is an essential regulator of DNA methylation that is highly expressed in many cancers. Here, we use transgenic zebrafish, cultured cells, and human tumors to demonstrate that UHRF1 is an oncogene. UHRF1 overexpression in zebrafish hepatocytes destabilizes and delocalizes Dnmt1 and causes DNA hypomethylation and Tp53-mediated senescence. Hepatocellular carcinoma (HCC) emerges when senescence is bypassed. tp53 mutation both alleviates senescence and accelerates tumor onset. Human HCCs recapitulate this paradigm, as UHRF1 overexpression defines a subclass of aggressive HCCs characterized by genomic instability, TP53 mutation, and abrogation of the TP53-mediated senescence program. We propose that UHRF1 overexpression is a mechanism underlying DNA hypomethylation in cancer cells and that senescence is a primary means of restricting tumorigenesis due to epigenetic disruption.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


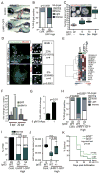
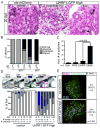
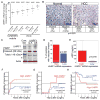
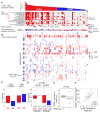
Similar articles
-
The Ubiquitin-like with PHD and Ring Finger Domains 1 (UHRF1)/DNA Methyltransferase 1 (DNMT1) Axis Is a Primary Regulator of Cell Senescence.J Biol Chem. 2017 Mar 3;292(9):3729-3739. doi: 10.1074/jbc.M116.750539. Epub 2017 Jan 18. J Biol Chem. 2017. PMID: 28100769 Free PMC article.
-
UHRF1 is an oncogene that promotes DNA hypomethylation.Cancer Discov. 2014 Apr;4(4):OF9. doi: 10.1158/2159-8290.CD-RW2014-036. Epub 2014 Feb 13. Cancer Discov. 2014. PMID: 24706678 No abstract available.
-
The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma.Mol Carcinog. 2016 Feb;55(2):209-19. doi: 10.1002/mc.22270. Epub 2015 Jan 16. Mol Carcinog. 2016. PMID: 25641194
-
[Role of UHRF1 in methylation regulation and angiogenesis].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2020 Feb 10;37(2):200-204. doi: 10.3760/cma.j.issn.1003-9406.2020.02.025. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2020. PMID: 32034755 Review. Chinese.
-
Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma.World J Gastroenterol. 2015 May 28;21(20):6317-28. doi: 10.3748/wjg.v21.i20.6317. World J Gastroenterol. 2015. PMID: 26034368 Free PMC article. Review.
Cited by
-
Genetic Studies on Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2022;1389:111-136. doi: 10.1007/978-3-031-11454-0_5. Adv Exp Med Biol. 2022. PMID: 36350508 Free PMC article.
-
Epigenomic and Other Evidence for Cannabis-Induced Aging Contextualized in a Synthetic Epidemiologic Overview of Cannabinoid-Related Teratogenesis and Cannabinoid-Related Carcinogenesis.Int J Environ Res Public Health. 2022 Dec 13;19(24):16721. doi: 10.3390/ijerph192416721. Int J Environ Res Public Health. 2022. PMID: 36554603 Free PMC article. Review.
-
The Mechanism Underlying the ncRNA Dysregulation Pattern in Hepatocellular Carcinoma and Its Tumor Microenvironment.Front Immunol. 2022 Feb 23;13:847728. doi: 10.3389/fimmu.2022.847728. eCollection 2022. Front Immunol. 2022. PMID: 35281015 Free PMC article. Review.
-
Natural and Synthetic Anticancer Epidrugs Targeting the Epigenetic Integrator UHRF1.Molecules. 2023 Aug 10;28(16):5997. doi: 10.3390/molecules28165997. Molecules. 2023. PMID: 37630248 Free PMC article. Review.
-
The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome.Genes (Basel). 2019 Feb 23;10(2):172. doi: 10.3390/genes10020172. Genes (Basel). 2019. PMID: 30813436 Free PMC article. Review.
References
-
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. - PubMed
-
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. - PubMed
-
- Babbio F, Pistore C, Curti L, Castiglioni I, Kunderfranco P, Brino L, Oudet P, Seiler R, Thalman GN, Roggero E, et al. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene 2012 - PubMed
-
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 1R01DK099558/DK/NIDDK NIH HHS/United States
- T32CA078207-14/CA/NCI NIH HHS/United States
- 5R01DK080789-02/DK/NIDDK NIH HHS/United States
- R01 DK076986/DK/NIDDK NIH HHS/United States
- R21 HG007394/HG/NHGRI NIH HHS/United States
- T32 CA078207/CA/NCI NIH HHS/United States
- 1R01DK076986/DK/NIDDK NIH HHS/United States
- R01 DK080789/DK/NIDDK NIH HHS/United States
- R01 DK099558/DK/NIDDK NIH HHS/United States
- F30DK094503/DK/NIDDK NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- 5P30CA006516-45/CA/NCI NIH HHS/United States
- F30 DK094503/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

