Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing
- PMID: 24487277
- PMCID: PMC4636053
- DOI: 10.1038/ng.2891
Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing
Abstract
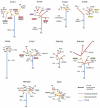
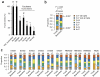
Clear cell renal carcinomas (ccRCCs) can display intratumor heterogeneity (ITH). We applied multiregion exome sequencing (M-seq) to resolve the genetic architecture and evolutionary histories of ten ccRCCs. Ultra-deep sequencing identified ITH in all cases. We found that 73-75% of identified ccRCC driver aberrations were subclonal, confounding estimates of driver mutation prevalence. ITH increased with the number of biopsies analyzed, without evidence of saturation in most tumors. Chromosome 3p loss and VHL aberrations were the only ubiquitous events. The proportion of C>T transitions at CpG sites increased during tumor progression. M-seq permits the temporal resolution of ccRCC evolution and refines mutational signatures occurring during tumor development.
Figures



Comment in
-
Kidney cancer: Intratumoral differences analysed.Nat Rev Urol. 2014 Mar;11(3):127. doi: 10.1038/nrurol.2014.43. Epub 2014 Feb 25. Nat Rev Urol. 2014. PMID: 24567088 No abstract available.
References
-
- Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. - PubMed
-
- Thirlwell C, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–1454. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

