Multi-algorithm and multi-model based drug target prediction and web server
- PMID: 24487966
- PMCID: PMC4647888
- DOI: 10.1038/aps.2013.153
Multi-algorithm and multi-model based drug target prediction and web server
Abstract
Aim: To develop a reliable computational approach for predicting potential drug targets based merely on protein sequence.
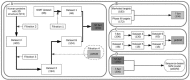
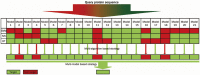
Methods: With drug target and non-target datasets prepared and 3 classification algorithms (Support Vector Machine, Neural Network and Decision Tree), a multi-algorithm and multi-model based strategy was employed for constructing models to predict potential drug targets.
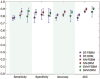
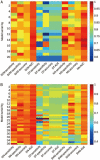
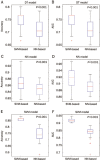
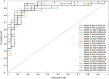
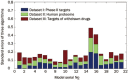
Results: Twenty one prediction models for each of the 3 algorithms were successfully developed. Our evaluation results showed that ∼30% of human proteins were potential drug targets, and ∼40% of putative targets for the drugs undergoing phase II clinical trials were probably non-targets. A public web server named D3TPredictor (http://www.d3pharma.com/d3tpredictor) was constructed to provide easy access.
Conclusion: Reliable and robust drug target prediction based on protein sequences is achieved using the multi-algorithm and multi-model strategy.
Figures


References
-
- 1Ohlstein EH, Ruffolo RR, Elliott JD. Drug discovery in the next millennium. Annu Rev Pharmacol Toxicol 2000; 40: 177–91. - PubMed
-
- 2Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov 2002; 1: 727–30. - PubMed
-
- 3Drews J. Drug discovery: a historical perspective. Science 2000; 287: 1960–4. - PubMed
-
- 5Drews J. Genomic sciences and the medicine of tomorrow. Nat Biotechnol 1996; 14: 1516–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

