Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types
- PMID: 24488081
- PMCID: PMC3951205
- DOI: 10.1038/ncomms4231
Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types
Abstract
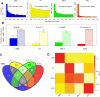
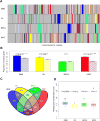
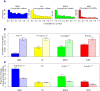
Prognostic genes are key molecules informative for cancer prognosis and treatment. Previous studies have focused on the properties of individual prognostic genes, but have lacked a global view of their system-level properties. Here we examined their properties in gene co-expression networks for four cancer types using data from 'The Cancer Genome Atlas'. We found that prognostic mRNA genes tend not to be hub genes (genes with an extremely high connectivity), and this pattern is unique to the corresponding cancer-type-specific network. In contrast, the prognostic genes are enriched in modules (a group of highly interconnected genes), especially in module genes conserved across different cancer co-expression networks. The target genes of prognostic miRNA genes show similar patterns. We identified the modules enriched in various prognostic genes, some of which show cross-tumour conservation. Given the cancer types surveyed, our study presents a view of emergent properties of prognostic genes.
Conflict of interest statement
Figures

References
-
- Sotiriou C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. - PubMed
-
- Bullinger L, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. New England Journal of Medicine. 2004;350:1605–1616. - PubMed
-
- Spentzos D, et al. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22:4700–10. - PubMed
-
- Adler AS, Chang HY. From description to causality: mechanisms of gene expression signatures in cancer. Cell Cycle. 2006;5:1148–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

