Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly
- PMID: 24488317
- PMCID: PMC3993358
- DOI: 10.1128/JB.01363-13
Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly
Abstract
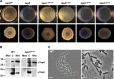
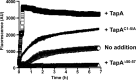
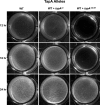
Bacillus subtilis biofilm formation relies on the assembly of a fibrous scaffold formed by the protein TasA. TasA polymerizes into highly stable fibers with biochemical and morphological features of functional amyloids. Previously, we showed that assembly of TasA fibers requires the auxiliary protein TapA. In this study, we investigated the roles of TapA sequences from the C-terminal and N-terminal ends and TapA cysteine residues in its ability to promote the assembly of TasA amyloid-like fibers. We found that the cysteine residues are not essential for the formation of TasA fibers, as their replacement by alanine residues resulted in only minor defects in biofilm formation. Mutating sequences in the C-terminal half had no effect on biofilm formation. However, we identified a sequence of 8 amino acids in the N terminus that is key for TasA fiber formation. Strains expressing TapA lacking these 8 residues were completely defective in biofilm formation. In addition, this TapA mutant protein exhibited a dominant negative effect on TasA fiber formation. Even in the presence of wild-type TapA, the mutant protein inhibited fiber assembly in vitro and delayed biofilm formation in vivo. We propose that this 8-residue sequence is crucial for the formation of amyloid-like fibers on the cell surface, perhaps by mediating the interaction between TapA or TapA and TasA molecules.
Figures



Similar articles
-
Tapping into the biofilm: insights into assembly and disassembly of a novel amyloid fibre in Bacillus subtilis.Mol Microbiol. 2011 Jun;80(5):1133-6. doi: 10.1111/j.1365-2958.2011.07666.x. Epub 2011 Apr 25. Mol Microbiol. 2011. PMID: 21488983
-
The majority of the matrix protein TapA is dispensable for Bacillus subtilis colony biofilm architecture.Mol Microbiol. 2020 Dec;114(6):920-933. doi: 10.1111/mmi.14559. Epub 2020 Jun 21. Mol Microbiol. 2020. PMID: 32491277
-
Mechanistic Insights into the Antibiofilm Activity of Simvastatin and Lovastatin against Bacillus subtilis.Mol Pharm. 2025 May 5;22(5):2703-2722. doi: 10.1021/acs.molpharmaceut.5c00191. Epub 2025 Mar 18. Mol Pharm. 2025. PMID: 40100146
-
The Role of Functional Amyloids in Multicellular Growth and Development of Gram-Positive Bacteria.Biomolecules. 2017 Aug 7;7(3):60. doi: 10.3390/biom7030060. Biomolecules. 2017. PMID: 28783117 Free PMC article. Review.
-
Functional amyloids in bacteria.Int Microbiol. 2014 Jun;17(2):65-73. doi: 10.2436/20.1501.01.208. Int Microbiol. 2014. PMID: 26418850 Review.
Cited by
-
Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum.Biomolecules. 2019 Nov 4;9(11):694. doi: 10.3390/biom9110694. Biomolecules. 2019. PMID: 31690032 Free PMC article.
-
Functional amyloids from bacterial biofilms - structural properties and interaction partners.Chem Sci. 2022 May 6;13(22):6457-6477. doi: 10.1039/d2sc00645f. eCollection 2022 Jun 7. Chem Sci. 2022. PMID: 35756505 Free PMC article. Review.
-
Defining the Expression, Production, and Signaling Roles of Specialized Metabolites during Bacillus subtilis Differentiation.J Bacteriol. 2021 Oct 25;203(22):e0033721. doi: 10.1128/JB.00337-21. Epub 2021 Aug 30. J Bacteriol. 2021. PMID: 34460312 Free PMC article.
-
Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms.Mol Microbiol. 2014 Aug;93(4):587-98. doi: 10.1111/mmi.12697. Epub 2014 Jul 18. Mol Microbiol. 2014. PMID: 24988880 Free PMC article. Review.
-
Bacillus subtilis EpsA-O: A novel exopolysaccharide structure acting as an efficient adhesive in biofilms.NPJ Biofilms Microbiomes. 2024 Oct 2;10(1):98. doi: 10.1038/s41522-024-00555-z. NPJ Biofilms Microbiomes. 2024. PMID: 39358392 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

