The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum
- PMID: 24488961
- PMCID: PMC3963592
- DOI: 10.1105/tpc.113.119255
The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum
Abstract
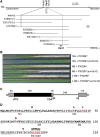
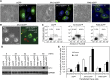
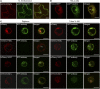
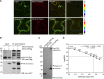
The recognition between disease resistance (R) genes in plants and their cognate avirulence (Avr) genes in pathogens can produce a hypersensitive response of localized programmed cell death. However, our knowledge of the early signaling events of the R gene-mediated hypersensitive response in plants remains limited. Here, we report the cloning and characterization of Xa10, a transcription activator-like (TAL) effector-dependent R gene for resistance to bacterial blight in rice (Oryza sativa). Xa10 contains a binding element for the TAL effector AvrXa10 (EBEAvrXa10) in its promoter, and AvrXa10 specifically induces Xa10 expression. Expression of Xa10 induces programmed cell death in rice, Nicotiana benthamiana, and mammalian HeLa cells. The Xa10 gene product XA10 localizes as hexamers in the endoplasmic reticulum (ER) and is associated with ER Ca(2+) depletion in plant and HeLa cells. XA10 variants that abolish programmed cell death and ER Ca(2+) depletion in N. benthamiana and HeLa cells also abolish disease resistance in rice. We propose that XA10 is an inducible, intrinsic terminator protein that triggers programmed cell death by a conserved mechanism involving disruption of the ER and cellular Ca(2+) homeostasis.
Figures



Similar articles
-
Induction of Xa10-like Genes in Rice Cultivar Nipponbare Confers Disease Resistance to Rice Bacterial Blight.Mol Plant Microbe Interact. 2017 Jun;30(6):466-477. doi: 10.1094/MPMI-11-16-0229-R. Epub 2017 Apr 25. Mol Plant Microbe Interact. 2017. PMID: 28304228
-
TAL effector-dependent Bax gene expression in transgenic rice confers disease resistance to Xanthomonas oryzae pv. oryzae.Transgenic Res. 2022 Feb;31(1):119-130. doi: 10.1007/s11248-021-00290-7. Epub 2021 Nov 8. Transgenic Res. 2022. PMID: 34748132
-
Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae.Plant Biotechnol J. 2015 Sep;13(7):993-1001. doi: 10.1111/pbi.12342. Epub 2015 Feb 3. Plant Biotechnol J. 2015. PMID: 25644581
-
The pepper Bs4C proteins are localized to the endoplasmic reticulum (ER) membrane and confer disease resistance to bacterial blight in transgenic rice.Mol Plant Pathol. 2018 Mar 30;19(8):2025-35. doi: 10.1111/mpp.12684. Online ahead of print. Mol Plant Pathol. 2018. PMID: 29603592 Free PMC article.
-
TAL effectors and the executor R genes.Front Plant Sci. 2015 Aug 20;6:641. doi: 10.3389/fpls.2015.00641. eCollection 2015. Front Plant Sci. 2015. PMID: 26347759 Free PMC article. Review.
Cited by
-
Identification of Bacterial Blight Resistance Loci in Rice (Oryza sativa L.) against Diverse Xoo Thai Strains by Genome-Wide Association Study.Plants (Basel). 2021 Mar 10;10(3):518. doi: 10.3390/plants10030518. Plants (Basel). 2021. PMID: 33802191 Free PMC article.
-
Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.).Theor Appl Genet. 2015 Oct;128(10):1933-43. doi: 10.1007/s00122-015-2557-2. Epub 2015 Jun 17. Theor Appl Genet. 2015. PMID: 26081948
-
Resistance Genes and their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)-an Updated Review.Rice (N Y). 2020 Jan 8;13(1):3. doi: 10.1186/s12284-019-0358-y. Rice (N Y). 2020. PMID: 31915945 Free PMC article. Review.
-
Transcriptional Reprogramming of Rice Cells by Xanthomonas oryzae TALEs.Front Plant Sci. 2019 Feb 25;10:162. doi: 10.3389/fpls.2019.00162. eCollection 2019. Front Plant Sci. 2019. PMID: 30858855 Free PMC article.
-
Available cloned genes and markers for genetic improvement of biotic stress resistance in rice.Front Plant Sci. 2023 Sep 5;14:1247014. doi: 10.3389/fpls.2023.1247014. eCollection 2023. Front Plant Sci. 2023. PMID: 37731986 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

