Type I IFN induces binding of STAT1 to Bcl6: divergent roles of STAT family transcription factors in the T follicular helper cell genetic program
- PMID: 24489092
- PMCID: PMC3967131
- DOI: 10.4049/jimmunol.1300675
Type I IFN induces binding of STAT1 to Bcl6: divergent roles of STAT family transcription factors in the T follicular helper cell genetic program
Abstract
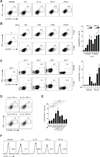
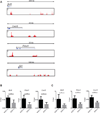
CD4(+) T follicular helper cells (TFH) are critical for the formation and function of B cell responses to infection or immunization, but also play an important role in autoimmunity. The factors that contribute to the differentiation of this helper cell subset are incompletely understood, although several cytokines including IL-6, IL-21, and IL-12 can promote TFH cell formation. Yet, none of these factors, nor their downstream cognate STATs, have emerged as nonredundant, essential drivers of TFH cells. This suggests a model in which multiple factors can contribute to the phenotypic characteristics of TFH cells. Because type I IFNs are often generated in immune responses, we set out to investigate whether these factors are relevant to TFH cell differentiation. Type I IFNs promote Th1 responses, thus one possibility was these factors antagonized TFH-expressed genes. However, we show that type I IFNs (IFN-α/β) induced B cell lymphoma 6 (Bcl6) expression, the master regulator transcription factor for TFH cells, and CXCR5 and programmed cell death-1 (encoded by Pdcd1), key surface molecules expressed by TFH cells. In contrast, type I IFNs failed to induce IL-21, the signature cytokine for TFH cells. The induction of Bcl6 was regulated directly by STAT1, which bound to the Bcl6, Cxcr5, and Pdcd1 loci. These data suggest that type I IFNs (IFN-α/β) and STAT1 can contribute to some features of TFH cells but are inadequate in inducing complete programming of this subset.
Figures



References
-
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. - PubMed
-
- Miller JF, De Burgh PM, Grant GA. Thymus and the production of antibody-plaque-forming cells. Nature. 1965;208:1332–1334. - PubMed
-
- Friedman H. Absence of Antibody Plaque Forming Cells in Spleens of Thymectomized Mice Immunized with Sheep Erythrocytes. Proc Soc Exp Biol Med. 1965;118:1176–1180. - PubMed
-
- Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

